USH2A SNPs: Difference between revisions
Tomemerald (talk | contribs) |
Tomemerald (talk | contribs) |
||
| Line 125: | Line 125: | ||
=== Functional significance === | === Functional significance === | ||
Here human individuals homozygous for S3743T could be examined for early loss of hearing accompanied by initial loss of mid-periferal and night vision. | Here human individuals homozygous for S3743T could be examined for early loss of hearing accompanied by initial loss of mid-periferal and night vision. They need not be homozygous because compound mutations would suffice, that is, a different USH2A mutation on the other chr1 allele. | ||
Alternatively, since mouse has an | Alternatively, since mouse has an reasonably conserved orthologous fibronectin domain, the effect of S3743N could be considered as a knockin. Here, even if the mouse gene did serve as a disease model for other alleles, symptoms for S3743N might or might not develop within the 2-3 year lifespan of laboratory mouse. | ||
For the immediate term, comparative genomics is best | For the immediate term, comparative genomics is best available guide. Here it is clear that S3743 is immensely conserved over several billion years of evolutionary time in those clades observable via genome projects (transcripts are too rare in this long gene to sample species diversity further). This establishes that N3743 is not part of the acceptable reduced alphabet at this residue, though T3743 at one time appears to have been acceptable in the teleost ancestor (and indeed is retained to the present day in early diverging deuterostomes). | ||
The difference alignment below of the exon containing S3743 shows overall conservation well above human proteome average but not extraordinary inflexibility at most positions. The fibronectin portion is evolving as well, no doubt through both drift, internal adaptive change, and co-evolutionary response to binding partner change. | |||
The large number of known loci throughout this protein that give rise to Usher Syndrome 2A suggest that not only does this protein play an exceedingly important structural role highly sensitive to mutation | Consequently S3743N -- despite its innocuous appearing nature (ie high Dayhoff matrix score) is likely to have significant non-adaptive impacts on either standalone structure of USH2A protein or its interaction with other proteins in the basement membrane. If selective pressure did not exist to maintain S3743, then what would account for its constancy despite copious variation in nearby residues over the same time spans? | ||
The large number of known loci throughout this protein that give rise to Usher Syndrome 2A suggest that not only does this protein play an exceedingly important structural role highly sensitive to seemingly minor mutation perturbations but also that no other gene product is able to compensate for its absence. | |||
=== Comparative genomics === | === Comparative genomics === | ||
Revision as of 13:19, 27 July 2009
USH2A
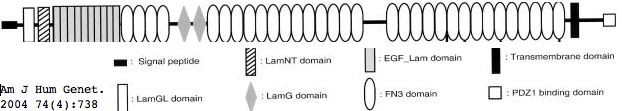
Usherin (USH2A), a 71-exon coding gene located on human chromosome 1q41], encodes a 5202 residue multi-domain protein comprised of a signal peptide, a PDZ1 binding domain (for USH1C and WHRN), 1 laminin NT-terminal domain, 10 laminin EGF-like domains, 4 fibronectin type-III domains (for collagen IV and fibronectin), and 2 laminin G-like domains followed by 31 additional fibronectin type-III domains all tethered to the cytoplasmic exterior by a single transmembrane domain.
The usherin gene is expressed in the basement membrane of many (but not all) cell types, notably in ear interstereocilia ankles and below retinal pigment epithelial cells (Bruch's layer). When normal function is disrupted by mutations in both copies, non-vestibular sensorineural deafness and degeneration of retinal photoreceptor cells called Usher syndrome type IIA results.
Initially, only the first 21 exons were studied but later it emerged that the gene was much longer and mutations along the entire length of the protein all led to the same disease: 125, 163, 230, 268, 303, 334, 346, 352, 478, 536, 595, 644, 713, 759, 1212, 1349, 1486, 1572, 1665, 1757, 2080, 2086, 2106, 2169, 2238, 2265, 2266, 2292, 2562, 2875, 2886, 3088, 3099, 3115, 3124, 3144, 3199, 3411, 3504, 3521, 3590, 3835, 3868, 3893, 4054, 4115, 4232, 4433, 4439, 4487, 4592, 4624, 4795, 5031.
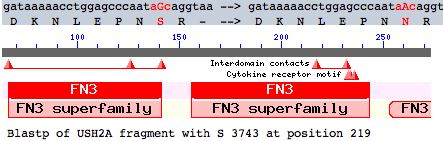
This note evaluates a tentative new SNP in USH2A with comparative genomics. The mutation occurs as a non-hotspot G-->A transition causing a seemingly innoculous S-->N amino acid change at postion 3743. This is just downstream from a glycosylation motif and very near known FN3 interdomain contact residues and a cytokine receptor motif (according to its annotation at SwissProt). This residue lies in the 22nd fibronectin domain which is split across exon 56 and 57.
This change will be shown significant (not plausibly neutral). It could represent an adaptive innovation but is more likely deleterious. The gene is single-copy so there are no prospects for compensation by a second gene. Consequently the mutation, if present on both alleles, could well result in a new form of Usher syndrome type IIA.
Background
USH2A, while resembling just another domain scramble, actually traces back to pre-blaterans in a coherent manner. Nearly full-length ortholog candidates can readily be recovered from sea anemone and hydra. Despite good representation in cnidarians (where its normal function may be similar), the gene seems completely lost (or majjorly diverged) from all arthropods and lophotrochozoans. It's not clear whether the need for a basement membrane organizer has been lost or whether some other gene has taken over USH2A's role in these clades.
Focusing now on the fibronectin FN3 domains (that envelop S3743), these are an ancient and exceedingly common domain in bilaterans with 2% of the human proteome containing them (400 genes), often in multiple tandem copies having a role in cell adhesion. However they are not particularly well conserved in primary sequence, though the tertiary structure likely holds up well enough for the structure at residue 3743 to be determined with both serine and asparagine present.
Here the best blastp match elsewhere within the human proteome to the FN3 domain containg residue 3743 is a fibronectin domain of PTPRQ, a dimly related protein tyrosine phosphatase with merely 28% of the fibronectin residues matching.
Internally, the best match to the other 30 FN3 domains of USH2A is not noticably better, suggesting very substantial divergence since these domains duplicated from a common source (either as internal tandems or domain shuffles). If as suggested above, USH2A had already assumed its contemporary domain structure in pre-bilateran metazoa, ample time has passed to produce the observed divergences between the individual domains.
While comparative genomics of intron positions and phases in a 71-exon protein are tedious to curationally pursue, the fibronectin domain containing S3743 falls across parts of two exons, the first of which is phase 21. This suggests that subsequent to the ancient intronation era, simple internal tandem duplications might not result in either a coherent reading phase or domain. Thus the domain structure of USH2A, while appearing somewhat arbitrary in its FN3 multiplicities, actually may be quite constrained by intronation against both contraction or expansion (in addition to whatever individual functional domain constrains exist).
As can be seen below, the internal fibronectin repeats are most often T threonine at the position corresponding to S3743 though other residues, not including the asparagine of S3743N, also occur. Here the numbering of better matches within the full length protein indicates they do not always correspond in match quality order to the linear order of the FN3 repeat within the protein.
FBN22 1 WSLPEKPNGLVSQYQLSRNGNLLFLGGSEEQNFTDKNLEPNS
WSLPEKPNGLVSQYQLSRNGNLLFLGGSEEQNFTDKNLEPNS
FBN.. 3702 WSLPEKPNGLVSQYQLSRNGNLLFLGGSEEQNFTDKNLEPNS
FBN22 1 WSLPEKPNGLVSQYQLSRNGN-LLFLGGSEEQNFTDKNLEPNS
WS+PEK NG++ +YQ+ + G L+ ++ + T L+P +
FBN.. 3610 WSVPEKSNGVIKEYQIRQVGKGLIHTDTTDRRQHTVTGLQPYT
FBN22 1 WSLPEKPNGLVSQYQLSRNGNLL-FLGGSEEQNFTDKNLEPNS
W PE+ NG++ Y+L RN L F N+TD+ L P S
FBN.. 4285 WIPPEQSNGIIQSYRLQRNEMLYPFSFDPVTFNYTDEELLPFS
FBN22 1 WSLPEKPNGLVSQYQLSRNGNLLFLGGSEEQNFTDKNLEPNS
W P K NG+++ Y + +G L N T +L P +
FBN.. 2553 WQHPRKSNGVITHYNIYLHGRLYLRTPGNVTNCTVMHLHPYT
FBN22 1 WSLPEKPNGLVSQYQLSRNGNLLFLGGSEEQNFTDKNLEPNS
W P PNG + Y+L R+G +++ G E + D L P
FBN.. 4464 WKPPRNPNGQIRSYELRRDGTIVYTG--LETRYRDFTLTPGV
FBN22 1 WSLPEKPNGLVSQYQLSRNGNLLFLGGSEEQNFTDKNLEPNS
W+ P+K NG+++QY L +G L++ G E+N+T +L +
FBN.. 2075 WNPPKKANGIITQYCLYMDGRLIYSG--SEENYTVTDLAVFT
FBN22 1 WSLPEKPNGLVSQYQLSRNGNLLFLGGSEEQNFTDKN-LEPNS
W P + NG + Y L RNG F G S +F+DK ++P
FBN.. 3521 WRKPIQSNGPIIYYILLRNGIERFRGTS--LSFSDKEGIQPFQ
FBN22 1 WSLPEKPNGLVSQYQLSRNGNLLFLGGSEEQNFTDKNLEPNS
W+ P PNG+V++Y + N L G + +F ++L P +
FBN.. 3040 WTSPSNPNGVVTEYSIYVNNKLYKTGMNVPGSFILRDLSPFT
FBN22 1 WSLPEKPNGLVSQYQLSRN-------GNLLFLGGSEEQNFTDKN--LEPNS
W P PNGLV + + R L+ L S F DK L P +
FBN.. 2644 WQPPTHPNGLVENFTIERRVKGKEEVTTLVTLPRSHSMRFIDKTSALSPWT
FBN22 1 WSLPEKPNGLVSQYQLSRNGNLLFLGGSEEQNFTDKNLEPNS
WS P + NG++ Y + +G L + G + + F + L+P +
FBN.. 4087 WSEPMRTNGVIKTYNIFSDGFLEYSGLNRQ--FLFRRLDPFT
FBN22 1 WSLPEKPNGLVSQYQLSRNGNLLFLGGSEE----QNFTDKNLEPNS
WS P+ PN Y L R+G ++ + Q F D +L P +
FBN.. 1074 WSPPDSPNAHWLTYSLLRDGFEIYTTEDQYPYSIQYFLDTDLLPYT
FBN22 1 WSLPEKPNGLVSQYQLSRNG------NLLFLGGSEEQNFTDK--NLEPNS
W PEKPNG++ Y + R ++LF+ F D+ L P +
FBN.. 3887 WMPPEKPNGIIINYFIYRRPAGIEEESVLFVWSEGALEFMDEGDTLRPFT
FBN22 1 WSLPEKPNGLVSQYQLSRNGNLLFLGGSEEQNFTDKNLEPNS
WS P NG +++Y L R N L G + +L+P S
FBN.. 4376 WSPPTVQNGKITKY-LVRYDNKESLAG-QGLCLLVSHLQPYS
FBN22 1 WSLPEKPNGLVSQYQL--------SRNGNLLFLGGSEEQNFTDKNLEPNS
W+ P +PNG V Y+L R N + + +F D L P +
FBN.. 4657 WTGPLQPNGKVLYYELYRRQIATQPRKSNPVLIYNGSSTSFIDSELLPFT
FBN22 1 WSLPEKPNGLVSQYQL------SRNGNLLFLGGSEE----QNFTDKNLEPNS
W P + NG + Y L R ++ + + Q++ L+P
FBN.. 4552 WDPPVRTNGDIINYTLFIRELFERETKIIHINTTHNSFGMQSYIVNQLKPFH
FBN22 1 WSLPEKPNGLVSQYQLSRN-------GN--------LLFLGGSEEQN---FTDKNLEPNS 42
WS P PNG + +Y++ R GN ++F + E+N + D L+P +
FBN.. 4175 WSEPVNPNGKIIRYEVIRRCFEGKAWGNQTIQADEKIVFTEYNTERNTFMYNDTGLQPWT 4234
Pseudogene issues
Long isoform USH2A transcripts are over 15,000 bp in length. Consequently position 3743 is not even represented in the set of all human direct transcripts. Even should a retrogene arise from retropositioing, it is unlikely that the process would extent upstream so many exons. Unsurprisingly no processed pseudogenes are evident in any mammalian genome (tblastn of wgs division of GenBank). Thus no potential for confusion exists in locating orthologs of USH2A even in distant species with incomplete genomes.
Paralog issues
No close paralog exists in the human proteome according to the UCSC GeneSorter track. The nearest matches are to other proteins containing laminin or fibronectin domains. No potential for confusion with other genes exists within vertebrates; however comparative genomics at and before teleost fish divergence needs more careful treatment because of whole genome and domain expansion.
Tandem domain repeat issues
In proteins with multiple copies of a given domain, both expansion and contraction can occur over evolutionary timescales resulting in different numbers of repeats in different clades. Under these circumstances it can be difficult to establish orthologs of a given domain. However here the fibronectin domains diverged early on and the 22nd domain seems to be present in all vertebrates with genome projects as a single-copy domain (meaning here no recent duplications or losses).
Known variations
There are no known issues with alternative splicing that would affect the fibronectin domain under consideration here. As noted earlier, a short version of the protein studied initially does not contain residue 3743 at all.
Structural significance
The 3D structure of the 22nd FN3 domain could be evaluated using best-blastp to a structurally determined FN3 domain in PDB, then modelling the FN3 domain in question by submitting it to SwissModel with both S3743 and T3743. Here the percent identity to an already-determined structure is mediocre but perhaps still sufficient.
If the serine at 3743 is on the surface and involved in a binding interaction with a second (unknown) protein, then the effect of the 3743N substitution would be very difficult to evaluate because asparagine and serine are of similar bulk and polarity. While S <--> N is a benign substitution at many positions in many proteins, at residue 3743 it appears that the hydroxyl is critical because, to the extent that any subsition at all is tolerated, it is threonine. (Bulk too may play a role because tyrosine is never seen.)
>pdb|1X5L|A Related structures Chain A, Solution Structure Of The Second Fn3 Domain Of Eph Receptor
Identities = 27/93 (29%), Positives = 44/93 (47%), Gaps = 16/93 (17%)
Query GVWVTPRHIIINSTTVELYWSLPEKPNGLVSQYQLSRNGNLLFLGGSEEQNF----------TDKNLEPNSRYTYKLEVKTGGG
V V R T+V L W PE+PNG++ +Y++ + E Q++ T L+P +RY +++ +T G
Sbjct QV-VVIRQERAGQTSVSLLWQEPEQPNGIILEYEIK-----YYEKDKEMQSYSTLKAVTTRATVSGLKPGTRYVFQVRARTSAG
Functional significance
Here human individuals homozygous for S3743T could be examined for early loss of hearing accompanied by initial loss of mid-periferal and night vision. They need not be homozygous because compound mutations would suffice, that is, a different USH2A mutation on the other chr1 allele.
Alternatively, since mouse has an reasonably conserved orthologous fibronectin domain, the effect of S3743N could be considered as a knockin. Here, even if the mouse gene did serve as a disease model for other alleles, symptoms for S3743N might or might not develop within the 2-3 year lifespan of laboratory mouse.
For the immediate term, comparative genomics is best available guide. Here it is clear that S3743 is immensely conserved over several billion years of evolutionary time in those clades observable via genome projects (transcripts are too rare in this long gene to sample species diversity further). This establishes that N3743 is not part of the acceptable reduced alphabet at this residue, though T3743 at one time appears to have been acceptable in the teleost ancestor (and indeed is retained to the present day in early diverging deuterostomes).
The difference alignment below of the exon containing S3743 shows overall conservation well above human proteome average but not extraordinary inflexibility at most positions. The fibronectin portion is evolving as well, no doubt through both drift, internal adaptive change, and co-evolutionary response to binding partner change.
Consequently S3743N -- despite its innocuous appearing nature (ie high Dayhoff matrix score) is likely to have significant non-adaptive impacts on either standalone structure of USH2A protein or its interaction with other proteins in the basement membrane. If selective pressure did not exist to maintain S3743, then what would account for its constancy despite copious variation in nearby residues over the same time spans?
The large number of known loci throughout this protein that give rise to Usher Syndrome 2A suggest that not only does this protein play an exceedingly important structural role highly sensitive to seemingly minor mutation perturbations but also that no other gene product is able to compensate for its absence.
Comparative genomics
............................................................^. USH2A_homSap GVWVTPRHIIINSTTVELYWSLPEKPNGLVSQYQLSRNGNLLFLGGSEEQNFTDKNLEPNSR USH2A_panTro GVWVTPRHIIINSTTVELYWSLPEKPNGLVSQYQLSRNGNLLFLGGSEEQNFTDKNLEPNSR USH2A_gorGor GVWVTPRHIIINSTTVELYWSLPEKPNGLISQYQLSRNGNLLFLGGSEEQNFTDKNLEPNSR USH2A_ponAbe GVWVTPRHIIINSTTVELYWSLPEKPNGLISQYQLSRNGNLLFLGGSEKQNFTDKNLEPNSR USH2A_nomLeu GVWVTPRHIIINSTTVELYWSLPEKPNGLISQYQLSRNGNLLFLGGSEEQKFTDKNLEPNSR USH2A_macMul GVWVTPRHIIINSTTVELYWSLPEKPNGLISQYQLSRNGNLLFLGGSEEQNFTDKNLEPNSR USH2A_calJac GVWVTPRHIIINSTTVELYWSLPEKPNGLISQYQLSRNGNLLFLGGSEEQNFTDKNLQPNSR USH2A_tarSyr GVWVTPRHIIINSTTVELYWSPPEKPNGLISQYQLSRNGTLLFLGGSEEQNFTDKNLEPHSR USH2A_micMur GVWVTPRHIIINSTTVELYWSPPEKPNGLISQYQLRRNGTLLFLGGSEEQNFTDKNLEPNSR USH2A_tupBel GVWVTPRHIIINSTTVELYWSLPKKPNGLISQYQLSRNGTLLFLGGSEEQNFTDKNLEPDSR USH2A_musMus GVWVTPRHIIINSTTVELYWNPPERPNGLISQYQLRRNGSLLLVGGRDNQSFTDSNLEPGSR USH2A_ratNor GVWVTPRHIIINSTTVELYWNPPERPNGVISQYRLRRNGSLLLVGGRDDQSFTDKNLEPNSR USH2A_dipOrd GVWVTPRHIIINSTAVELYWSPPEKPNGLISQYQLSRNGSVLFLGGREEQMFTDTNLEPNSR USH2A_cavPor GVWVTPRHTVINSTSVELYWSPPEKPNGLISQYRLSRNGTLLFVGGGEEQNFTDKHLEPNSR USH2A_speTri GVWVTPRHMIINSTTVELYWSPPEKPNGLISQYQLSRNGTLLLLGGSEERNFTDKHLEPNSR USH2A_oryCun GVWVTPRHIIINSTTVELYWTPPEKPNGLISQYQLNRNGIVVFLGGSKEQNFTDRNLKPNSR USH2A_ochPri GVWVSPRHIVINCTAVILYWSPPEKPNGIISQYQLIRNETVLYLGSGKEQNFTDGNLEPNSR USH2A_vicPac GVWVTPRHIIINPTTVELYWSPPEKPNGLISQYQLSRNGTLVFLGGSEEQNFTDKNLEPNSR USH2A_susScr GVWVTPRHIIVNSTTVELYWSLPEKPNGLISQYQLSRNGTVVFLGGSEERNFTDKNLEPNSR USH2A_turTru GVWVTPRHIIINSTTVELYWSLPEKPNGLISQYQLSRNGSLVFLGGSEEQNFTDKNLEPNSR USH2A_bosTau GVWVTPRHIVVNSTTVELFWSPPEKPNGLVSQYQLSRNGSLIFLGGSEEHNFTDKNLEPNSR USH2A_equCab GVWMTPRHIIINSTTVELYWSPPENPNGLISQYQLSRNGTLVFLGGSEEQNFTDKNLEPNSR USH2A_felCat GVWVTPRHIIINSTTVELYWSPPEKPNGLISQYQLSRNGTLVFLGGNEEQNFTDKNLEPNSR USH2A_canFam GVWVTPRHIIINSTTVELYWNPPEKPNGLISQYQLSRNGTLVFLGGSEEQNFTDKNLEPNSR USH2A_myoLuc GVWATPRHIIINATAVELYWRPPERPNGLISRYQLIRNGTSVFLGGSEDQHFTDHNLAPNSR USH2A_pteVam GVWVTPQHIIINSTAVELCWSPPEEPNGLISQYRLSRDGNLVFLAGAEEHCFTDKNLEPNSR USH2A_loxAfr GVWLTPRHIIINPTTVELYWSQPEKPNGLISRYHLRRNGTLVLLGGSEEQNFTDKNLEPNSR USH2A_proCap GVWMTPRHIVINSTTVELHWSLPEKPNGHISQYRLRRNGTLVFQGGGEEQNFTDTNLEPNSR USH2A_dasNov GVWVTPGHIIINSTTVELYWSQPEKPNGLISHYQLSRNGTLIFLAGREEQSFTDKNLEPNSR USH2A_choHof GVWVTPQHIIINSTTVELYWSQPEKPNGLISQYQLSRNGTSVFQGGREEQHFTDKNLEPSSR USH2A_monDom GVWSIPRHIIINSTTVELYWNEPEKPNGLISKYQLHRNGTVIFLGGREDQNFTDDSLEPKSS USH2A_ornAna GVWSKPQHITVSSTTVELYWSQPEKPNGVISQYRLIRNGTEIFAGTRDSLNFTDDSLESNSR USH2A_galGal GVWPKPHHIIVSSTEVEIYWSEPEIPNGLITQYRLFRDEEQIFLGGSRDLNFTDVNLQPNSR USH2A_taeGut GVWPKPHHIIVSSTEVEMYWSEPEEPNGLITHYRLFRDGEQIFLGGSTARNFTDVNLQPNSR USH2A_anoCar GVWSQPRHVIVSSKIVELYWDEPEEPNGIISLYRLFRNGEEIFMGGELNLNFTD-TVQPNNR 4 traces, not in assembly USH2A_xenTro GVWSNPYHVTINESVLELYWSEPETPNGIVSQYRLILNGEVISLRSGECLNFTDVGLQPNSR USH2A_tetNig GVWSKPRHLTVNASAVELHWDPPQQPQGLVSQYRLKRDGRAVFTGDHLQRNYTDAGLQPQRR USH2A_takRub GVWSKPRHLIVTTAVVELYWDPPQQPHGHISQYKLKRDGQTVFTGDHDDQNYTDTGLRPHRR USH2A_gasAcu GVWSSPRHVVINTSAVELYWDQPLQPNGHISQYRLNRDGDTIFTGDHREQNYTDTGLLPNRR USH2A_oryLat GVWSKPRHLIINTSAVELYWDQPSQPNGLISQYRLIRDGLTVFTGARRDQNYTDTGLEPKRR USH2A_danRer GVWSMPRHIQLNSSAVELHWSDPLKLNGLLSGYRLLRDGELVFTADGGKMSYTDAGLQPNTR USH2A_calMil GIWPKPCHVIVNSSTVELYWTEPEKPNGIITQFRLLRDNAVIYTGTRRNRNYTDAGLQPDTR USH2A_braFlo QEVSRPRFVVVSSTEIEVYWSEPGRPNGIITQYQLVRDGSVIYSGG--DMNFTDSGLTPSTT XM_002214612 aligns over 2807 aa USH2A_strPur EGLMQPTHVVVSSTILELYWFEPSQPNGVITSYILYRDDELVYSGNNSVLTYVDTGLTPNTR XM_788345 aligns over 5030 aa USH2A_nemVec SQQPAPVITVSSSRRLDLAWSPPDNPNGIILRYELYRNGTEVYRG--VIRGYNDTNLQPDTL XM_001638773 aligns over 3005 aa USH2A_hydMag SQQGAPFVLFQTSRLINIGWFPPDNLNGILIKYELYRDRTKIFVG--LDNNYTDNNLKPYTY XM_002165140 USH_homSap GVWVTPRHIIINSTTVELYWSLPEKPNGLVSQYQLSRNGNLLFLGGSEEQNFTDKNLEPNSR USH_panTro .............................................................. USH_gorGor .............................I................................ USH_macMul .............................I................................ USH_calJac .............................I...........................Q.... USH_ponAbe .............................I..................K............. USH_nomLeu .............................I....................K........... USH_turTru .............................I.........S.V.................... USH_tupBel .......................K.....I.........T...................D.. USH_susScr ..........V..................I.........TVV.......R............ USH_tarSyr .....................P.......I.........T...................H.. USH_micMur .....................P.......I.....R...T...................... USH_vicPac ............P........P.......I.........T.V.................... USH_felCat .....................P.......I.........T.V....N............... USH_canFam ....................NP.......I.........T.V.................... USH_equCab ...M.................P..N....I.........T.V.................... USH_bosTau .........VV.......F..P.................S.I.......H............ USH_cavPor ........TV....S......P.......I...R.....T...V..G........H...... USH_speTri ........M............P.......I.........T..L......R.....H...... USH_oryCun ....................TP.......I.....N...IVV.....K......R..K.... USH_dipOrd ..............A......P.......I.........SV.....R...M...T....... USH_loxAfr ...L........P........Q.......I.R.H.R...T.VL................... USH_dasNov ......G..............Q.......I.H.......T.I..A.R...S........... USH_choHof ......Q..............Q.......I.........TSV.Q..R...H........S.. USH_proCap ...M.....V........H.........HI...R.R...T.V.Q..G.......T....... USH_musMus ....................NP..R....I.....R...S..LV..RDN.S...S....G.. USH_ratNor ....................NP..R...VI...R.R...S..LV..RDD.S........... USH_myoLuc ...A........A.A.....RP..R....I.R...I...TSV......D.H...H..A.... USH_pteVam ......Q.......A...C..P..E....I...R...D...V..A.A..HC........... USH_monDom ...SI...............NE.......I.K...H...TVI....R.D.....DS...K.S USH_ochPri ....S....V..C.A.I....P......II.....I..ETV.Y..SGK......G....... USH_ornAna ...SK.Q..TVS.........Q......VI...R.I...TEI.A.TRDSL....DS..S... USH_galGal ...PK.H...VS..E..I...E..I....IT..R.F.DEEQI.....RDL....V..Q.... USH_taeGut ...PK.H...VS..E..M...E..E....ITH.R.F.D.EQI.....TAR....V..Q.... USH_xenTro ...SN.Y.VT..ESVL.....E..T...I....R.IL..EVIS.RSG.CL....VG.Q.... USH_anoCar ...SQ...V.VS.KI.....DE..E...II.L.R.F...EEI.M..ELNL....-TVQ..N.