B-Cells
From genomewiki
B-Cells
- B-Cells recognize non-body stuff, which is called antigen. They do this by either binding to a part of the the antigen called epitope with receptors on their cell membrane. Or they can secrete these receptors in the form of antibodies into the blood (or mucus) where the antibodies will bind themselves to antigen, without any other cell around.

Schematic diagram of a typical antibody showing two Ig heavy chains (blue) linked by disulfide bonds to two Ig light chains (green). The constant (C) and variable (V) domains are shown.
- An Antibody comes in five different flavors (in placental mammals) IgA, IgD, IgE, IgG, IgM. They all share the same basic structure, but differ in their additions. Somehow we assume that IgG is the most important, because it is the most common type and we don't know that much about the other types anyways.
- IgG is one of those antibody types that looks like a Y. The *ends* of the arms of the Y recognize the antigen, the *ends* not the middle part.
- The two arms of the Y of all antibodies - and so IgG - are composed of two identical Heavy and two identical Light Chains
- The two ends of the Y, the part that recognizes the antigen are together called Fab. One Fab comprises a bit of the heavy chain and a bit of the light chain. Not everything touches the antigen, so the most important region in the Fab fragment is called Fv (variable).
- The Fv can be split into its three main parts, the complementary determining regions, called CDR1, CDR2 and CDR3. Remember that there is a light and a heavy chain in one Fv, so there are six complementary determining regions in total.
- Of the three heavy-chain CDRs, CDR3 has a much larger diversity than CDR1 and CDR2. Structural studies showed that CDR3 is the main antigen-binding site. It was believed that CDR1 and CDR2 interacted with the MHC molecule, while CDR3 interacted with the antigen. However, later CDR1 was also shown to have some contact with the antigen. CDR1 and CDR2 are easier to investigate (no hypermutation, see below)
- For a long time, no one knew how the body produces the right Fa fragments that bind to "non-self" but not to "self". A popular theory was that proteins "fold" themselves into the right shape, around some part of antigen (epitope). This turned out to be wrong, the answer lies in a particular property of the genome of these cells: Each B-cell has a different genome and each cell produces only one antibody.
B-cell genomes
- Apart from transposable elements, virus insertions and random mutations, all somatic cells (=not egg or sperm) have the same genome. There are only two types of cells that change their genome somatically: B-cells and T-cells.
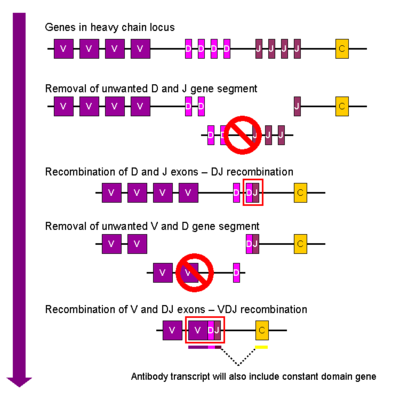
- B-cells are one of these two somatic cell types that can glue together two different parts of their genome (recombine), in what is called somatic recombination. This happens during embryonic development and only once. The loci that recombine are called IGH@ (heavy chains), IGK@ and IGL@ (K=kappa, L=lamba, both are light chains) (how the heck do you pronounce the @ here? NB: The @ is HGNC standard and means that the identifier is not a gene, but a locus of genes, e.g. HOX@, see this search). They are located on chromosomes 14 (heavy), 2 (kapp) and 22 (lambda). (NB: do not confuse IGH, the locus, with the antibody types IgA, IgE, IgM, IgD and IgG).
- On these loci many (somewhere between 6-60) little genes are located next to each other. From left to right, they are separated into a stretch of V genes, then D genes, then J genes, though light chain loci do not have D genes, so we usually write it as V(D)J. On the genome browser, these loci don't look great, because all VDJ genes are all very similar, so the RefSeq sequences match everywhere and are filtered out.
- All Vs, Ds and Js are separated by recombination signal sequences. The recombinase is activated for a short time during embryonic B-cell development (the mammals probably got that enzyme that from a transposase) and first recombines a D to J and then a V to this DJ combination.
- While this is going on, a terminaldeoxyneucleotidetransferase adds and deletes some nucleotides between the different segments to add a little bit of noise between them.
- The final sequence is often written as V-N1-D-N2-J, N1 and N2 being the little additions by the terminaldeoxyneucleotidetransferase
- While B cells divide, Somatic hypermutation will change random nucleotides within the full receptor sequence. This is different from the nucleotide additions, in that no inserts/deletes occur, only mutations
- The V segment is a lot longer than the D or J segments. Vs can be identified with a simple BLAST search, whereas Js and Ds are better recognizes with HMMs.
B-cell diversity
- As a result, each B-cell has its own unique combination of VDJ genes in all of their loci. That means that each B-cell is different from each other B-cell when we are very young.
- In each cell, this V(D)J-combination is transcribed to produce a heavy or light chain immunoglobulin protein and assembled into receptors
- At around 2 months of embryonic development, all B-cells that bind something in the body kill themselves. At our age, only B-cells circulate in our blood that bind to stuff that is *not* part of the body. All others have been eliminated already.
- In any of these B-cells, when the receptors built from the VDJ combinations bind to some antigen, this particular B-cell will start to divide and many of the daughter cells will produce antibodies from this VDJ combination. In addition, the daughter cells will hypermutate their VDJ segments and those that bind better will turn out to produce even more daughter cells and more antibodies. Macrophages will then kill the cells that are marked with antibodies.