Dating Doppel (PRND)
Dating doppel: missing from bird and fish genomes -- but present in marsupial, lizard and frog
The prion and doppel genes represent a local tandem duplication that has descended intact in most lineages with the two genes remaining adjacent and unpseudogenized (though both spawned retroposed pseudogenes in later lineages). When did this gene duplication take place, did it correlate with or facilitate an evolutionary advantage, and how have the (largely unknown) normal functions of the original gene since diverged? Retention of either copy is already somewhat puzzling in view of viability of PRNP knockouts, indeed PRNP/PRND double knockouts.
The strikingly weak homology between prion and doppel -- below 30% identity within-species even trimming to conserved core -- suggests either an ancient duplication (lamprey era?) followed by symmetric divergence at more or less average rates, or alternatively a recent duplication (tetrapod era?) followed by rapid asymmetrically fast evolution on the doppel side as it neofunctionalized or subfunctionalized to a secondary function of the unduplicated gene. These possibilities can be distinguished by looking at earlier diverging species that bracket date on the duplication event.
If the duplication is old, then doppel should be found in various draft vertebrate genomes now available -- 34 mammals (see Genome completion status) plus chicken, lizard, frog, zebrafish, tetraodon, fugu, medaka, and stickleback. If synteny (gene order and orientation) are conserved, doppel will lie between PRNP and RASSF2, allowing highly diverged forms to be recognized by blast focused to that intergene region that otherwise might be lost among whole-genome match debris). Synteny and weak blast support are hopefully supplemented by conservation of internal signatures such as signal peptide, post-translationally modified arginine, disulfides, glycosylation sites, alpha helices, short beta sheet (YMLG in human), and GPI attachment.
Repeat structure is trickier to use because it has been totally lost in all PRND (at the time of duplication?) and in PRNP varies from wildly expanded in fish, biphasic hexapeptide in frog, turtles, birds, and lizards to decapeptide (platypus), nonapeptide (marsupial), or octapeptide (placentals) as discussed elsewhere Prion repeat evolution. Further, the number of observed repeats varies markedly across individuals (sometimes pathologically), an effect seen from birds to mammals. It is an unstable coding region of very high GC subject to frequent replication slippage with new variants constantly arising. There is no wild type allele, only a choice within population variants. This requires reconstructed ancestral nodes to consist of a reconstructed population. Consequently the region is not strictly speaking homologous across species and is only inappropriately included in orthologous alignments. The normal function if any is unknown; it cannot be assumed that a single function holds across species (knockins have no complementation assay). Thus the distribution of functions across PRNP and PRND must address possibly non-uniform function in PRNP across species.
PRND may never have been able to fully complement PRNP. Not only has the repeat region been lost (most parsimoniously at, or shortly after, duplication), but the two genes are asymmetrically positioned with respect to upstream promoters and regulatory elements of the original single gene, ie PRND is in the rain shadow of PRNP. After considerations involving dating and removing subsequently accrued retroposons, upstream non-coding elements were too far removed from the PRNP start codon in the ancestral genome to have been included in the original duplication. PRND must then have been transcribed by read-through (as seen today in rare mouse transcripts) which would have given it appropriate developmental and tissue expression from the get-go, perhaps at a reduced level that mitigated gene dosage while allowing more rapid divergence. In this asymmetric scenario, PRND only later developed transcription control regions in the intergenic region that could separately modulate expression.
Tandem gene duplications are thus quite different from large segmental (or whole genome) duplications. They require further stratification depending on whether parallel or inverted, whether inclusive or not of upstream regulatory regions, and whether or not in comparable euchromatin environments. Subsequent history is also important: PRNP and PRND have remained largely unperturbed but other tandem gene duplications have been followed by much later (thus lineage-specific) secondary events such as further tandem expansion, separating block inversions, translocation of one copy elsewhere, and deletion. Such events accrue roughly in proportion to elapsed time.
Consequently older tandem gene duplications tend to have unique histories, undercutting any program to develop sweeping theories. Rate histories must speak to a single timeline (of reconstructed ancestral nodes leading to a fixed species) rather than to repeatedly bifurcated timelines of phylogenetic tree data that reference increasingly incomparable clades. The history of the PRNP/PRND tandem duplication below will illustrates that.
PRNP sequences are available at GenBank from over 100 species of placental mammal. Dozens more can be extracted from genome projects in various stages of completion. The lesser effort expended on PRND and its much rarer transcripts conspire with rapidly weakening percent identity to make advanced bioninformatic techniques necessary as pipeline annotation collapses.
The brushtail opossum (Trichosurus vulpecular) prion gene sequence was determined in 1995. It conserves structural elements overall but presents various anomalies such as nonapeptide repeats. Its sequence can be used as query at the UCSC genome browser to locate a second complete marsupial PRNP sequence from Monodelphis domestica. Monodelphis domestica PRND is located next to PRNP in the same orientation in a gapless region of the assembly, left- and right-flanked by syntenically appropriate genes. A third marsupial sequence (Macropus eugenii) is available from unassembled NCBI trace archives; full length PRNP and PRND sequences are readily covered there by trace blastn using opossum query, though synteny cannot be pursued. Marsupials have had longer to diverge than placental mammals; the two 'opossums' here are not closely related, Didelphimorphia vs Diprotodontia. Paired rate studies thus await PRND from Trichosurus vulpecular and a fossil calibration of an overall molecular clock for marsupial divergence.
Monotreme represents an important earlier divergence. PRNP and PRND can be recovered from the newly released assembly of the platypus genome, Ornithorhynchus anatinus. This species, which diverged after birds but before the Theran split, has a PRNP decapeptide x 4 repeat (rather than a bird- or turtle-like hexapeptide. However its sequence, PQGGGASWGH, is unusual for its glutamine in second position and terminal histidine (reverse of usual mammal residues). In other respects it is conventional. Unfortunately neither sequence is available for echidna; these could be easily acquired because both genes consist of a single short coding exon.
Within placental mammals, doppel homology runs at about 85% identity but this drops off to 48% relative to marsupial and 41% in platypus, indicating (relative to global proteomes) a fast-evolving protein not yet a pseudogene in any lineage (though it spawned 'processed' pseudogenes in kangaroo rat and stem xenarthra). Indeed, with the density of PRNP gene sequences in placentals and the mammalian phylogenetic tree topology finally determined, clade-specific rate variations could be pursued in unusual detail, along with any correlated doppel evolution. The ancestral boreoeuthere PRND and PRNP sequences provided below could facilitate that quest by providing non-roundtrip rates over a fixed 85 myr time interval. The ancestral sequences have a meagre 21% identity.
The chicken genome is gap-free in this area and retains mammalian gene order and orientation (RASSF2- PRNP+ SCL23A-). However, no dopple gene (nor evidence for a decayed pseudogene) occurs in chicken in the expected position despite various high-sensitivity blast probes (reconstructed ancestral doppel, anole PRND). If prion gene duplication occurred prior to chicken divergence, then doppel has decayed to an unrecognizable pseudogene, translocated to region of gappy coverage, or deleted. The other bird genome underway, Taeniopygia guttata (finch), also lacks PRND. The intergenic region where PRND should lie, between PRNP and RASSF2, consists in chicken of a meagre 529 bp vs 122,057 bp in human, establishing a large indel has occured.
Very recently, a lizard genome became available, Anolis carolinensis. Lizards are a sister group to birds. Using specialized methods, a full length PRND gene can be located in expected syntenic position and validated by retained internal signatures despite an exceedingly very weak blast match.
Similarly the diploid frog, Xenopus tropicalis, has an unmistakable doppel gene at expected syntenic position and orientation. Some sequence uncertainty exists because of assembly issues and frameshifts in the two supporting ESTs. No doppel can be found in the (non-genomic) tetraploid frog, Xenopus laevis, though it has two actively transcribed PRNP genes that differ slightly in the repeat-like region as well at a few individual amino acids. There may be two in-paralog doppels in this species, depending on the relative timing of PRNP duplication and the tetraploidization event..
These results have a simple parsimonious explanation: the PRND duplication predated amphibian divergence, it was retained in all lineages except birds, where a lineage-specific deletion occured bird clade. Crocodile sequencing of this region could further refine the date of this event.
However nothing prevents homoplasy of tandem duplications and indeed we shall see this in teleost fish. On the contrary, the observation that both copies of a given duplication can be useful over billions of year of branch length supports the likelihood of it happening again in a lineage not participating in the original duplication. Here lizard and frog doppel queries retrieve solely mammalian and turtle PRND genes when back-blasted against the GenBank non-redundant nucleotide division. Both lack the repeat region and retain some internal diagnostic doppel signatures though not the second disulfide bond. This argues against the idea of multiple independent duplications of the prion gene in lizard and frog distinct from a later lineage-specific duplication in mammals (with birds then never having a PRND to delete).
The prion gene in teleost fish has radically diverged from its mammalian counterpart especially in the repeat region but remains recognizable via various conserved internal features, residual synteny, and its weak alignment. Fugu has a second PRNP gene on a different chromosome (presumably resulting from the whole genome duplication in Teleostei) plus an apparent novel tandem duplication that resembles PRNP rather than PRND. Coelocanth genome has been proposed but not begun -- that could further refine the timing of tetrapod PRND duplication.
As it stands, that event occurred after teleost fish divergence but before that of amphibians, say 400 million years ago. The prion gene clearly represents the ancestral form and presumably ancestral function, ie, a doppel-like ancestor did not duplicate to a second gene that become PRNP. No counterpart of PRNP can be located in the initial release of the chondrichthyian genome, Callorhinchus milii, but that will soon be finished to higher coverage and regional assembly. PRNP is most likely an ancient gene that simply has become unrecognizable.
Doppels are evolving noticeably faster than prions but have not become pseudogenes in any species investigated to date. (Very recent pseudogenes that have not yet acquired internal stop codons or loss of deeply invariant residues are difficult to detect.) It must be remembered that PRNP's globular region lies within the most slowly evolving quartile of mammalian genes. As a representative example, human/dog proteins have 78% identity for PRND but 87% for PRNP. A reconstructed ancestral mammal sequence for PRND shows no sign whatsoever of converging to a PRNP-like sequence, percent identity ancester-to-ancestor remains in the mid-20's. Since this uses up 100 million years of the 400 available, doppel may have evolved more drastically at an earlier time, perhaps soon after the duplication event.
The most economical explanation of the data is that the slow evolving prion gene retains the lion's share of ancestral functions whereas doppel has a narrow new but possibly related function with limited and specialized expression in uncommon cell types, yet one important enough for longterm gene retention. Over longer evolutionary time spans, we can't be sure either gene really functions identically in all species.
To further investigate the co-evolution of PRNP and PRND, it is imperative to obtain additional pairs of gene sequences from a phylogenetically representative sample of species, which here means echidna, turtles, and more lizards and amphibians. PRNP tends to be over-sampled within Artiodactylia, especially Bovidae. Doppel remains very poorly represented at dbEST even in much-studied mammals with enriched libraries of tissues like testis where doppel is known to be transcribed. Consequently additional doppel sequences will not drop out as byproducts of transcriptome projects.
However a respectable number of doppel sequences have accumulated. In addition to genome projects, van Rheede and coworkers sequenced nine doppels from important early diverging clades of placental mammals. PRND from the chimpanzee genome has special interest for evaluating known human doppel polymorphisms to determine "wildtype" human. Macaque doppel also exhibits the 2 amino acid deletion seen in human at position 122-123, showing this event predated old world monkey divergence. In fact that event can be precisely timed to the stem lying between tarsier and new world monkey divergence and shown highly non-homoplasic.
Ancestral length and PRND indels
Here an interesting loss of 2 residues helps define primates from new world monkeys on. Other indels in this region have limited phylogenetic interest because of homoplasy -- support for a hedgehog and shrew sister relation to the exclusion of other laurasiatheres is undercut by similar deletion in kangaroo rat, mouse lemur and pika.
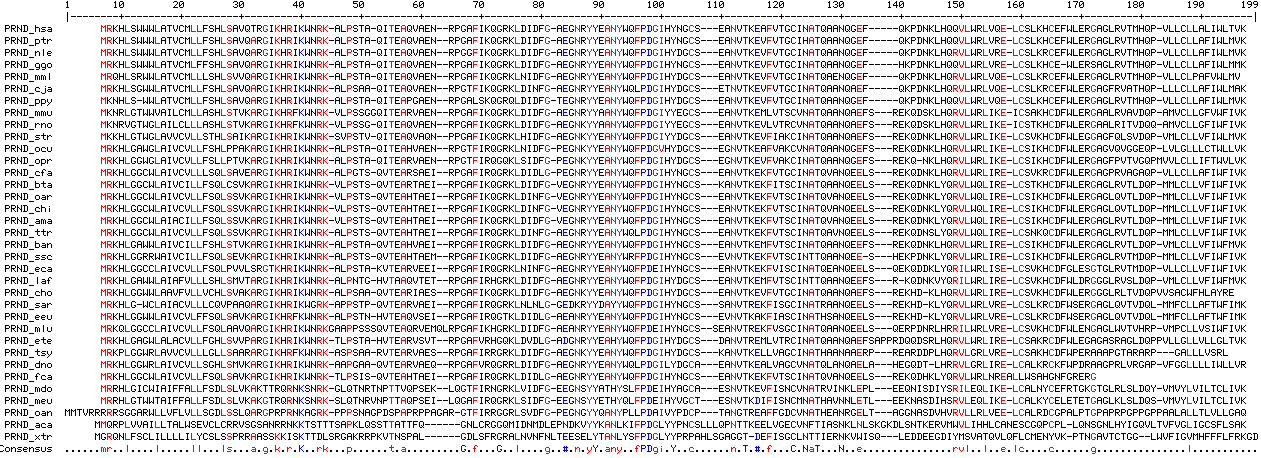
prnd_hsa anqgefq--kpdnklhqqvlwrlvqelcs homo sapiens (human) prnd_ptr anqgefq--kpdnklhqqvlwrlvrelcs pan troglodytes (chimp) prnd_ggo anqgefh--kpdnklhqqvlwrlvrelcs gorilla gorilla (gorilla) prnd_ppy anqgefq--kpdnklhqqvlwrlvqelcs pongo pygmaeus (orang_sumatran) prnd_nle anqgefq--kpdnklhqrvlwrlvrelcs nomascus leucogenys (gibbon) prnd_mml enqgefq--kpdnklhqrvlwrlvqelcs macaca mulatta (rhesus) prnd_pha anqgefq--kpdnklhqrvlwrlvqelcs papio hamadryas (baboon) prnd_cja anqaefq--kpdnklhqrvlwrlvqelcs callithrix jacchus (marmoset_nwm) prnd_tsy anaaerprearddplhqrvlgrlvrelcs tarsius syrichta (tarsier) prnd_oga anqaefsrekqdnklhqrilwrlirelcs otoleumur garnetti (bushbaby) prnd_mmr anqaefarekqd-klherilwrltrelcs microcebus murinus (mouse_lemur) prnd_tbe anqaefskekqdnklyqrvlwrlikelcs tupaia belangeri (tree_shrew) prnd_ocu anqgefsrekqdnklhqrvlwrlikelcs oryctolagus cuniculus (rabbit) prnd_opr anqgefsrekq-nklhqrvlwrlikelcs ochotona princeps (pika) prnd_dor anqaefaremqd-kfyqrvlwrltkelca dipodomys ordii (kangaroo_rat) prnd_str anqaefsrekqdnklhqrvlwrlikelcs spermophilus tridecemlineatus (ground_squirrel) prnd_rno anqaefsrekqdsklhqrvlwrlikeics rattus norvegicus (rat) prnd_mmu anqaefsrekqdsklhqrvlwrlikeics mus musculus (mouse) prnd_cpo anqaefsrekqdnklhqrilwrlikelcs cavia porcellus (guinea_pig) prnd_sar anqeelsrekhd-klyqrvlwrlvrelcs sorex araneus (shrew) prnd_eeu anqeelsrekhd-klyqrvlwrlvrelcs erinaceus europaeus (hedgehog) prnd_cfa anqeelsrekqdnklhqrvlwrlirelcs canis familiaris (dog) prnd_fca anqeelsrekqddklyqrvlwrlnr-ecs felis catus (cat) prnd_tte enqeelsrekqddklhqrilwrlirelcs tapirus terrestris (tapir) prnd_eca anqeelsqekqddklyqrilwrliselcs equus caballus (horse) prnd_mlu anqeelsqerpdnrlhrrilwrlvrelcs myotis lucifugus (microbat) prnd_csp vnqeelsqekqdkllhqrilwqlirelcs cynopterus sphinx (bat) prnd_oar anqeelsrekqdnklyqrvlwqlirelcs ovis aries (sheep) prnd_chi anqeelsrekqdnklyqrvlwqlirelcs capra hircus (goat) prnd_bta anqeelsrekqdnklyqrvlwqlirelcs bos taurus (cow) prnd_ttr vnqeelsrekqdnslyqrvlwqlirelcs tursiops truncatus (dolphin) prnd_ssc anqee-shekpdnklyqrvlwrlirelcs sus scrofa (pig) prnd_pcw vnqeelslekpdnklyqrvlwrlirelcs physeter catodon (whale) prnd_ban anqeefsrekqdnklhqrvlwrlirelcs boreoeuthere ancestralis (boreoeuthere) prnd_dno anqaelaherqd-tlhgrvlgrlirelca dasypus novemcinctus (armadillo) prnd_cho anqaefsrekhd-klhqrvlwrlirelcs choloepus hoffmanni (sloth) prnd_pcp vnqeefsrekqdnklyqrilwrlirelcs procavia capensis (hyrax) prnd_ete anqaefsapqqdsrlhqrvlwrlirelcs echinops telfairi (tenrec) prnd_ema anqeefsrekqdnkvyqrilwrlirelcs elephas maximus (elephant) prnd_laf anqeefsr-kqdnkvyqrilwrlirelcs loxodonta africana (elephant) prnd_tma anqeefsrekqdnklyqrilwrlirelcs trichechus manatus (manatee) prnd_mdo inklepleeqnisdiysrileqlikelca monodelphis domestica (opossum) prnd_meu vnnletleeknasdihsrvleqlikelca macropus eugenii (wallaby) prnd_oan anrgeltaggnasdvhvrvllrlveelca ornithorhynchus anatinus (platypus) prnd_aca snklnlskgkdlsntkervmwvlihhlca anolis carolinensis (lizard) prnd_xtr rnkvwisqleddeegdiymsvatqvlqflcm xenopus tropicalis (frog)
Phylogenetic distribution of available PRND sequences
11 primates (euarchonta) 7 glires (rodents and rabbits) 16 laurasiatheres 1 ancestral boreoeuthere 2 xenarthrans 5 aftrotheres 2 marsupials 1 platypus 0 bird (gene deleted in chicken and finch) 1 lizard 1 amphibian
Reference collection of PRND sequences
Below 45 PRND doppel sequences from land vertebrates are provided. Most are complete and reliable, but others are fragmentary and no better than the ongoing genome project from which they were extracted (initially low coverage, perhaps a single applicable trace). The phylogenetic distribution is approximately half as extensive as for PRNP. Almost all species having an available PRND would also have an available PRNP. It would be very easy to extend PRND coverage since primer design presents no issues. The most interesting rare genomic event for PRND of any phylogenetic depth is the indel shown above.
>PRND_homSap Homo sapiens (human) full
MRKHLSWWWLATVCMLLFSHLSAVQTRGIKHRIKWNRKALPSTAQITEAQVAENRPGAFIKQGRKLDIDFGAEGNRYYEANYWQFPDGIHYNGCSEANVTKEAFVTGCINATQAANQGEFQKPDNKLHQQVLWRLVQELCSLKHCEFWLERGAGLRVTMHQPVLLCLLALIWLTVK*
>PRND_ptanTro Pan trogdylotes (chimpanzee) full
MRKHLSWWWLATVCMLLFSHLSAVQARGIKHRIKWNRKALPSTAQITEAQVAENRPGAFIKQGRKLDIDFGAEGNRYYEANYWQFPDGIHYNGCSEANVTKEVFVTGCIHATQAANQGEFQKPDNKLHQQVLWRLVRELCSLKHCEFWLERGAGLRVTMHQPVLLCLLAFIWLMVK*
>PRND_gorGor Gorilla gorilla (gorilla)
MRKHLSWWWLATVCMLFFSHLSAVQARGIKHRIKWNRKALPSTAQITEAQVAENRPGAFIKQGRKLDIDFGAEGNRYYEANYWQFPDGIHYNGCSEANVTKEVFVTGCINATQAANQGEFHKPDNKLHQQVLWRLVRELCSLKHCEWLERSAGLRVTMHQPVLLCLLAFIWLMMK*
>PRND_ponPyg Pongo pygmaeus (orangutan) shares 2 bp deletion unique to primates.
MKNHLSWWLATVCMLLFSHLSAVQARGIKHRIKWNRKALPSTAQITEAPGAENRPGALSkQGRQLDIDFGTEGNRYYEANYWQFPDGIHYDGCSEANVTKEVFVTGCINATQAANQGEFQKPDNKLHQQVLWRLVQELCSLKHCEFWLERGAGLRVTMHQPVLLCLLAFIWLMVK*
>PRND_nomLeu Nomascus leucogenys (gibbon)
MRKHLSWWWLATVCMLLFSHLSAVQARGIKHRIKWNRKALPSTAQITEAQVAENRPGGFIKQGRKLDIDFGAEGNRYYEANYWQFPDGIHYDGCSEANVTKEVFVTGCINATQAANQGEFQKPDNKLHQRVLWRLVRELCSLKRCEFWLERGAGLRVTMHQPVLLCLLAFIWLMVK*
>PRND_papHam Papio hamadryas (baboon)
MRKHLSWWWLATVCMLLFSHLSAVQTRGIKHRIKWNRKALPSTAQITEAQVAENRPGAFIKQGRKLDIDFGAEGNRYYEANYWQFPDGIHYNGCSEANVTKEAFVTGCINATQAANQGEFQKPDNKLHQQVLWRLVQELCSLKHCEFWLERGAGLRVTMHQPVLLCLLALIWLTVK*
>PRND_macMul Macaca mulata (macaque) full
MRQHLSRWWLATVCMLLLSHLSVVQARGIKHRIKWNRKALPSTAQITEAQVAENRPGAFIKQGRKLNIDFGAEGNRYYEANYWQFPDGIHYDGCSEANVTKEVFVTGCINATQAENQGEFQKPDNKLHQRVLWRLVQELCSLKRCEFWLERGAGLRVTMHQPVLLCLPAFVWLMV*
>PRND_calJac Callithrix jacchus (marmoset) new world monkey 89%
MRKHLSGWWLATVCMLLFSHLSAVQARGIKHRIKWNRKALPSAAQITEAQVAENRPGTFIKQGRKLDINFGAEGNRYYEANYWQLPDGIHYDGCSETNVTKEVFVTGCINATQAANQAEFQKPDNKLHQRVLWRLVQELCSLKRCEFWLERGAGFRVATHQPLLLCLLAFIWLMAK*
>PRND_tarSyr Tarsius syrichta (tarsier)
MRKPLGGWRLAVVCVLLLGLLSAARARGIKHRFKWNRKASPSAARVTEARVAESRPGAFIRRGRKLDIDFGAEGNKYYEANYWQFPDGIHYDGCSKANVTKELLVAGCINATHAANAAERPREARDDPLHQRVLGRLVRELCSAKHCDFWPERAAAPGTARARPGALLLVSRL
>PRND_micMur Microcebus murinus (mouse_lemur)
MRKHLGGWRPAIVFILLFSHLAVVKARGIKHRIKWNRKPLPSTVHITEALVAENRPGAFIKQGRKLDVDFGEEGNRYYEANYWQFPDGIHYDGCSDANVTKEVFVASCINATVAANQAEFAREKQDKLHERILWRLTRELCSVKRCEFWLERGAGLRVAEDCPTILSLLLLLWLLLR*
>PRND_tupBel Tupaia belangeri tree shrew 6.7 m tr frag
RALPSTAHITEARVAENRPGAFIRQGRKLDIDFGVEGNRYYEANYWQFPDGIHYDGCSEVNVTKEMFITSCINATQAANQAEFSKEKQDNKLYQRVLWRLIKELCSIKHCDFWLERGAGLRVTMHLPVMLCLLVFVWFVVK*
>PRND_musMus Mus musculus (mouse) full
MKNRLGTWWVAILCMLLASHLSTVKARGIKHRFKWNRKVLPSSGGQITEARVAENRPGAFIKQGRKLDIDFGAEGNRYYAANYWQFPDGIYYEGCSEANVTKEMLVTSCVNATQAANQAEFSREKQDSKLHQRVLWRLIKEICSAKHCDFWLERGAALRVAVDQPAMVCLLGFVWFIVK*
>PRND_ratNor Rattus norvegicus (rat) full
MKNRVGTWGLAILCLLLASHLSTVKARGIKHRFKWNRKVLPSSGQITEAQVAENRPGAFIKQGRKLDIDFGAEGNKYYAANYWQFPDGIYYEGCSEANVTKEVLVTRCVNATQAANQAEFSREKQDSKLHQRVLWRLIKEICSTKHCDFWLERGAALRITVDQQAMVCLLGFIWFIVK*
>PRND_cavPor Cavia porcellus (guinea pig) fragment AY130773 N-extended at traces 7,499,469 seq
MKTSLCVWALVCVLLQCHFSSVAARGIKHRIRWGRKPTPSPSQVTEARVAMTRPGAFIKEGHKLNIDFGAEGNRYYETNYWQFPDGIHYNGCSDTNVTKEVLVTSCINATQAANQAEFSREKQDNKLHQRILWRLIKELCSA
>PRND_dipOrd Dipodomys ordii (squirrel) pseudogene
ITEARYRENAQGPSSIKP--QADIDF--ERAQ*YEA*YWQFADGINYEGCSRRHVTKQMFVSRCINVTQAANQAEFAREMQD-KFYQRVLWRLTKELCAVKQCDFWLERGEGCPVAIVNRPAMLGLLLCMWFIL
>PRND_speTri Spermophilus tridecemlineatus (squirrel) full
MKKHLGTWGLAVVCVLLSTHLSAIKARGIKHRIKWNRKSVPSTVQITEAQVAQNPPGAFIKQGRKLHIDFGAEGNKYYEANYWQFPDGIYYDGCSEANVTKEVFIAKCINATQAANQAEFSREKQDNKLHQRVLWRLIKELCSVKHCDFWLEGGAGFQLSVDQPVMLCLLVFIWLMVK*
>PRND_oryCun Oryctolagus cuniculus (rabbit) revised complete from traces 8,968,713 sequences 75%
MRKHLGAWGLAIVCVLLFSHLPPAKARGIKHRIKWNRKALPSTAQITEAHVAENRPGTFIRQGRKLNIDFGPEGNKYYEANYWQFPDGVHYDGCSEGNVTKEAFVAKCVNATQAANQGEFSREKQDNKLHQRVLWRLIKELCSIKHCDFWLERGAGVQVGGEQPLVLGLLLCTWLLVK*
>PRND_ochPri Ochotona princeps (pika) full
MRKHLGGWGLAIVCVLLFSLLPTVKARGIKHRIKWNRKALPSTAQITEARVAENRPGTFIRQGQKLSIDFGPEGNKYYEANYWQFPDGIHYDGCSEGNVTKEVFVAKCINATQAANQGEFSREKQNKLHQRVLWRLIKELCSIKHCDFWLERGAGFPVTVGQPMVVLCLLIFTWVLVK*
>PRND_canfam Canis familiaris (dog) full
MRKHLGGCWLAIVCVLLLSQLSAVEARGIKHRIKWNRKALPGTSQVTEARSAEIRPGAFIRQGRKLDIDLGPEGNRYYEANYWQFPDGIHYNGCSEANVTKEKFVTGCINATQVANQEELSREKQDNKLHQRVLWRLIRELCSVKRCDFWLERGAGPRVAGAQPVLLCLLAFIWFIVK*
>PRND_ailMel Ailuropoda melanoleuca (panda) full 89% identical dog
MRKHLGGCWLAIVCVLLFSQLSAVKARGIKHRIKWNRKALPSTSQVTEARTAEIRPGAFIRQGRKLDIDLGAEGNRYYEANYWQFPDGIHYNGCSEANVTKEKFVSGCINATRVANQEELSREKQDNRLYQRVLWRLVRELCSVKHCDFWLERGAGLRVGGDQALLLCLLVFIWFIVK*
>PRND_felCat Felis catus (cat) improved fragment 8,248,053 traces now 9.2
MRKHLGGCWLAIVCVLLFSQLSAVKARGIKHRIKWNRKTLPSISQVTEAHTAEIRPGAFIRQGRKLDIDLGAEGNRYYEANYWQFPDGIHYNGCSEANVTKEKFVTSCINATQVANQEELSREKQDhKLYQRVLWRLNREALLWSAHGNFGRERG
>PRND_equCab Equus caballus (horse)
MRKHLGGCCLAIVCVLLFSQLPVVLSRGTKHRIKWNRKALPSTAKVTEARVEEIRPGAFIRQGRKLNINFGAEGNRYYEANYWQFPDEIHYNGCSEANVTKEKFVISCINATQEANQEELSQEKQDDKLYQRILWRLISELCSVKHCDFGLESGTGLRVTMDQPVLLCLLVFIWFIVK*
>PRND_tapTer Tapirus terrestris (tapir) fragment
KALPSTSKVTEAHTAEIRPGAFIRQGRKLNIDFGAEGNRYYEANYWQFPDEIHYNGCSEANVTKEKFVISCINATQAENQEELSREKQDDKLHQRILWRLIRELCSV
>PRND_sorAra Sorex araneus (shrew) Laurasiatheria; Insectivora 7.23M tr
MRKHLGWCLAIACVLLLCQVPAAQARGIKHRIKWGRKAPPSTPQVTEARVAEIRPGAFIRQGRKLNLNLGGEDKRYYDAYYWQFPDGIHYNGCSSANVTREKFISGCINATRAANQEELSREKHDKLYQRVLWRLVRELCSLKRCDFWSERGAGLQVTVDQLMMFCLLAFTWFIMK*
>PRND_eriEur Erinaceus europaeus (hedgehog)10,952,642 seq complete
MRKHLGGWWLAIVCVLFFSQLSAVKARGIKHRFKWNRKALPSTNHVTEAQVSEIRPGAFIRQGTKLDIDLGAEANRYYEANYWQFPDGIHYNGCSEVNVTKAKFIASCINATHSANQEELSREKHDKLYQRVLWRLVRELCSLKRCDFWSERGAGLQVTVDQLMMFCLLAFTWFIMK*
>PRND_myoLuc Myotis lucifugus (microbat) full
MRKQLGGCCLAIVCVLLFSQLAAVQARGIKHRIKWNRKGAAPPSSSQVTEAQRVEMQLRPGAFIKHGRKLDIDFGAEANRYYEANYWQFPDEIHYNGCSSEANVTREKFVSGCINATQAANQEELSQERPDNRLHRRILWRLVRELCSVKHCDFWLENGAGLWVTVHRPVMPCLLVSIWFIVK*
>PRND_pteVam Pteropus vampyrus (macrobat) frameshifts frag
MRRQLGGCWLAIVCVLLFSQLSTVKARGIKHRTKWNR PCDRVS VTESQ GAFIKQGRK
>PRND_cynSph Cynopterus sphinx (bat) fragment
KTLPSPSSHVTESQTAEMRPGAFVKQGRKLNIDFGDEGNRYYEAHYWEFPDGIYYDACSKANVTKEKFVTSCINATQAVNQEELSQEKQDKLLHQRILWQLIRELCSV
>PRND_bosTau Bos taurus (cow) full
MRKHLGGCWLAIVCILLFSQLCSVKARGIKHRIKWNRKVLPSTSQVTEARTAEIRPGAFIKQGRKLDIDFGVEGNRYYEANYWQFPDGIHYNGCSKANVTKEKFITSCINATQAANQEELSREKQDNKLYQRVLWQLIRELCSTKHCDFWLERGAGLRVTLDQPMMLCLLVFIWFIVK*
>PRND_oviAri Ovis aries (sheep) full
MRKHLGGCWLAIVCVLLFSQLSSVKARGIKHRIKWNRKVLPSTSQVTEAHTAEIRPGAFIKQGRKLDINFGVEGNRYYEANYWQFPDGIHYNGCSEANVTKEKFVTSCINATQVANQEELSREKQDNKLYQRVLWQLIRELCSIKHCDFWLERGAGLQVTLDQPMMLCLLVFIWFIVK*
>PRND_capHir Capra hircus (goat)
MRKHLGGCWLAIVCVLLFSQLSSVKARGIKHRIKWNRKVLPSTSQVTEAHTAEIRPGAFIKQGRKLDINFGVEGNRYYEANYWQFPDGIHYNGCSEANVTKEKFVTSCINATQVANQEELSREKQDNKLYQRVLWQLIRELCSIKHCDFWLERGAGLQVTLDQPMMLCLLVFIWFIVK*
>PRND_conTau Connochaetes taurinus (wildebeest) EF192574
MRKHLGGCWLAIVCVLLFSHLSSVKARGIKHRIKWNRKVLPSTSQVTEARTAEIRPGAFIKQGRKLDIDFGVEGNRYYEANYWQFPDGIHYNGCSEPNVTKENFITSCINATQVANQEEMSREKQDNKLYQRVLWQLIRELCSIKHCDFWLERGAGLQVTLDQPMMLCLLVFIWFIVK*
>PRND_girCam Giraffa camelopardalis (giraffe) EF192572
MRKHLGRCWLAIVCVLLFSQLSSVKARGIKHRIKWNRKVLPSTSQVTEAHTAEIRPGAFIKQGRKLDIDFGVEGNRYYEANYWQFPDGIHYNGCSEANVTKEEFVTSCINATQAANQEELSREKQDNKLYQRILWRLIRELCSIKHCDFWLERGAGLQVTLDQPMMLCLLVFIWFIVK*
>PRND_ama Antidorcas marsupialis (springbok) Bovidae; Antilopinae
MRKHLGGCWLAIACILLFSQLSSVKARGIKHRIKWNRKVLPSTSQVTEAHTAEIRPGAFIKQGRKLDIDFGVEGNRYYEANYWQFPDGIHYNGCSEANVTKEKFVTSCINATQVANQEELSREKQDNKLYQRVLWQLIRELCSIKHCDFWLERGAGLRVTLDQPMMLCLLVFIWFIVK*
>PRND_turTru Tursiops truncatus (dolphin)
MRKHLGGCWLAIVCVLLFSQLSSVKARGIKHRIKWNRKALPSTSQVTEAHTAEIRPGAFIRQGRKLDINFGAEGNRYYEANYWQLPDGIHYNGCSEANVTKEKFVTSCINATQAVNQEELSREKQDNSLYQRVLWQLIRELCSNKQCDFWLERGAGLRVTLDQPMMLCLLVFIWFIVK*
>PRND_pseCra Pseudorca crassidens (false killer whale) AP009417
MRKHLGGCWLAIVCVLLFSQLSSVKARGIKHRIKWNRKALPSTSQVTEAHTAEIRPGAFIRQGRKLDINFGAEGNRYYEANYWQFPDGIHYNGCSEANVTKEKFVTSCINATQAVNQEELSREKQDNNLYQRVLWQLIRELCSNKQCDFWLERGAGLRVTLDQPMMLCLLVFIWFIVK*
>PRND_phyCat Physeter catodon (sperm) whale middle fragment
KALPSTSQVTEAHTAEIVPGAFIKQGRKLDIDFGAEGNRYYEANYWQFPDGIHYNGCSEASVTKEKFVTSCINATQEVNQEELSLEKPDNKLYQRVLWRLIRELCSI
>PRND_susScr Sus scrofa (pig) full length ti|470951185
MRKHLGGRRWAIVCILLFSQLSEVKARGIKHRIKWNRKALPSTSQVTEAHTAEMRPGAFIKQGRKLDIDFGAEGNRYYEANYWRFPDGIHYNGCSEVNVTKEKFVTSCINTTQAANQEESHEKPDNKLYQRVLWRLIRELCSIKHCDFWLERGAGLRVTMDQPMMLCLLVFIWLIVK*
>PRND_lamPac Lama pacos (alpaca) wgs frag
MRKHLGGCWLALACVLLLSPLSAVKARGIKHRIRWNRRPLPSASQATEAYTAELRPG
>PRND_borAnc Boreoeuthere ancestralis (ancestral)
MRKHLGAWWLAIVCILLFSHLSTVKARGIKHRIKWNRKALPSTAQVTEAHVAEIRPGAFIRQGRKLDIDFGAEGNRYYEANYWQFPDGIHYNGCSEANVTKEMFVTSCINATQAANQEEFSREKQDNKLHQRVLWRLIRELCSIKHCDFWLERGAGLRVTLDQPVMLCLLVFIWFMVK*
>PRND_dasNov Dasypus novemcinctus (armadillo) pseudo frag
ANYWQLPDGILYDGCAEANVTKEALVAGCVNGHTAANQAELAHERQDTLHGRVLGRLIRELCALKRCKFWPDR
>PRND_dasNov Dasypus novemcinctus (armadillo)
MRKHLGGWRLAIVCVLLSGHLSMVKARGIKHRIKWNRKAAPGAAQVTEARVAEQRPGAFVRQGRRLDIDFGAEGNRYYEANYWQLPDGILYDGCAEANVTKEALVAGCVNATQLANQAELAHEGQDTLHRRVLGRLIRELCALKRCKFWPDRAAGPRLVRGAPVFGGLLLLIWLLVR*
>PRND_choHof Choloepus hoffmanni (sloth) single trace old dasypus pseudogene, ti|563245674
MRKHLGGWWLAAVFVLLVCHLSVAKARGIKHRIKWNRKALPSAAQVTEARIAESRPGAFIKQGRKLDIDFGAEGNKYYEANYWQFPDGIHYDGCSEANVTKEVFVTGCINATQAANQAEFSREKHDKLHQRVLWRLIRELCSVKHCDFWLERGGGLRLTVDQPVVSACWFHLAYRE*
>PRND_losAfr Loxodonta africana (elephant) Afrotheria 176 aa revised/corrected
MRKHLGAWWLAIAFVLLLSHLSMVTARGIKHRIKWNRKALPNTGHVTAAQVTETRPGAFIRHGRKLDIDFGAEGNRYYEANYWQFPDGIHYDGCSEANVTKEMFVTSCINTTQAANQEEFSRKQDNKVYQRILWRLIRELCSVKHCDFWLDRGGGLRVSLDQPVMLCLLVFIWFMVK*
>PRND_eleMax Elephas maximus (elephant) Afrotheria fragment
KALPNTGHVTAAQVTETRPGAFIRHGRKLDIDFGAEGNRYYEANYWQFPDGIHYDGCSEANVTKEMFVTSCINTTQAANQEEFSREKQDNKVYQRILWRLIRELCSV
>PRND_proCap Procavia capensis (cape rock hyrax) Afrotheria fragment AY130772 ti|1293358170
KPVTNPAHMTAAQVSDRRLGTFIRHGRKLDIDFGAEGNRYYEANYWLFPDGIHYDGCSDTNVTKELFVTNCINTTQAVNQEEFSREKQDNKLYQRILWRLIRELCSllglDFWPERVGGLffSLDQPVMLCLLVFIWFMLK*
>PRND_echTel Echinops telfairi (tenrec) Afrotheria 10,323,158 seq
MRKHLGAGWLALACVLLFGHLSVVPARGIKHRIKWNRKTLPSTAHVTEARVSVTRPGAFVRHGQKLDVDLGADGNRYYEAHYWQFPDGIHYDGCSDANVTREMLVTRCINATQAANQAEFSAPPRDQQDSRLHQRVLWRLIRELCSAKRCDFWLEGAGASRAGLDQPPVLLGLLVLLGLTVK*
>PRND_triMan Trichechus manatus (manatee) Afrotheria fragment
KPLPNTPHVTAAQVSDARPGAFIRHGRKLDIDFGAEGNRYYEANYWQFPDGIHFDGCSEANVTKEMFVTSCINTTQAANQEEFSREKQDNKLYQRILWRLIRELCSV
>PRND_monDom Monodelphis domestica (opossum) doppel genomic revised +rassf2 -prnd -prnp
MRRHLGICWIAIFFALLFSDLSLVKAKTTRQRNKSNRKGLQTNRTNPTTVQPSEKLQGTFIRNGRKLVIDFGEEGNSYYATHYSLFPDEIHYAGCAESNVTKEVFISNCVNATRVINKLEPLEEQNISDIYSRILEQLIKELCALNYCEFRTGKGTGLRLSLDQYVMVYLVILTCLIVK*
>PRND_macEug Macropus eugenii (wallaby)
MRRHLGTWWTAIFFALLFSDLSLVKAKGTRQRNKSNRKSLQTNRVNPTTAQPSEILQGAFIRQGRKLSIDFGEEGNSYYETHYQLFPDEIHYVGCTESNVTKDIFISNCMNATHAVNNLETLEEKNASDIHSRVLEQLIKELCALKYCELETETGAGLKLSLDQSVMVYLVILTCLIVK*
>PRND_sacHar Sarcophilus harrisii (tasmanian_devil) single exon gene 77% macEug
MRTPLETWWIAIFFTLLFSDLSLVKAKGIRQRNKSNRKSLQTNRANPTREQPSKILQGTFIRKGRKLSINFGEEGNSYYEAHYKLFPDEIHYVGCAESSVTKDVFISNCVNVTHTANKLEPPEERNSSAIYSRVLEQLIKELCALKYCEFGMQIGAGFRLSLDQSMMVYLMILAFFIVK*
>PRND_ornana Ornithorhynchus anatinus (platypus) 42% to opposum 187 aa 4 cys in register
MMTVRRRRRSGGARWLLVFLVLLSGDLSSLQARGPRPRNKAGRKPPPSNAGPDSPAPRPPAGARGTFIRRGGRLSVDFGPEGNGYYQANYPLLPDAIVYPDCPTANGTREAFFGDCVNATHEANRGELTAGGNASDVHVRVLLRLVEELCALRDCGPALPTGPAPRPGPPGPPAALALLTLVLLGAQ*
>PRND_chrPic Chrysemys picta (painted_turtle)
MGRTRMVTCWMAMLLLVLCSNITLCRRGSSSKKASQNKSPPPVNKPSPPTKKEPVKIPGTLCYTGQLLDIKLEPEEAKYYTDNFKKFPDCIYYPRCFQSLEPNATKDTLVSECFNVTVSLTENKLDLPEGKNATSVYSRVMWQVISHLCAMEFCCQPCSLALSIHGSFGWLAMLCLGSFISLTMQ*
>PRND_anocar Anolis carolinensis (lizard) weak but real! scaffold_1221:78,884-117,121 syntenic, oriented like PRNP but no larger
MMQRPLVVAILLTALWSEVCLCRRVSGSANRRNKKTSTTTSAPKLQSSTTATTFQGNLCRGGQMIDNMDLEPNDKVYYKANLKIFPDGLYYPNCSLLLQPNTTKEELVGECVNFTIASNKLNLSKGKDLSNTKERVMWVLIHHLCANESCGQPCPLLQNSGNLHYIGQVLTVFVGLIGCSFLSAK*
>PRND_pytMol Python molurus (python) AEQU010389504
MWKNLLVVVLLAVLCSNVGLCRRRSGPTRKWPEKTVSKSSTKSSTTLGNLCYGSEMIDNMILEANDTTFYKDNAKEFPNGLFYPNCSLPLQPKVSRETRTEECVDFIIASSNLTLPNGKNASDPSKRVMWLVINHLCASETCSLPCPGSSNVSSAHYFLPILMTALVGYSILATR
>PRND_xenTro Xenopus tropicalis (frog) 182 aa single exon 2 cDNAs odd unbalanced cysteines syntenic turtle-prnp like, no laevis
MGRQNLFSCLILLLLILYCSLSSPRRAASSKKISKTTDLSRGAKRRPKVTNSPALGDLSFRGRALNVNFNLTEESELYTANLYSFPDGLYYPRPAHLSGAGGTDEFISGCLNTTIERNKVWISQLEDDEEGDIYMSVATQVLQFLCMENYVKPTNGAVTCTGGLWVFIGVMHFFFLFRKGD*
>PRND_anaCar Anolis carolinensis dna query sequence
atgatgcagagacctctggtagtagccatccttttgacagccctatggagcgaggtctgtctctgtcggcgagtgagcgg
atctgcaaataggcggaataagaagacttccaccaccacaagtgctcccaaacttcaaagctcaaccacagccacaacct
tccaaggaaatctttgtcgtggtggtcaaatgattgacaatatggacctcgagccaaatgacaaagtctattacaaggca
aacctgaagattttcccagatggactttattatcccaactgttccttgcttctgcagcccaacacgaccaaagaagagct
cgttggtgaatgcgtcaatttcaccattgcatcaaataagttgaacctatctaaaggcaaggatttgagcaacacaaagg
agagggtgatgtgggtcttgattcatcacttgtgtgcaaatgagtcgtgcggtcagccttgccctttgctccaaaactct
ggaaatctccactatattggccaagtactcactgtatttgttggcctaattggttgctcttttctttctgcaaaataa
* * * * ** *** * * * * * * * **
PRND_homSap -MRKHLSWWWLATVCMLLFSHLSAVQTRGIKHRIKWNRKALPSTA-QITEA-QVAENRPGAFIKQGRKLD-IDFGAEGNRYYEANYWQFPDGIHYNGCS---EANVTKEAFVTGCINATQAANQGEF--QKPDNKLHQQVLWRLVQELCSLKHCEFWLERGAGLRVTMHQPVL-LCLLALIWLTVK---
PRND_ptanTro -MRKHLSWWWLATVCMLLFSHLSAVQARGIKHRIKWNRKALPSTA-QITEA-QVAENRPGAFIKQGRKLD-IDFGAEGNRYYEANYWQFPDGIHYNGCS---EANVTKEVFVTGCIHATQAANQGEF--QKPDNKLHQQVLWRLVRELCSLKHCEFWLERGAGLRVTMHQPVL-LCLLAFIWLMVK---
PRND_gorGor -MRKHLSWWWLATVCMLFFSHLSAVQARGIKHRIKWNRKALPSTA-QITEA-QVAENRPGAFIKQGRKLD-IDFGAEGNRYYEANYWQFPDGIHYNGCS---EANVTKEVFVTGCINATQAANQGEF--HKPDNKLHQQVLWRLVRELCSLKHCE-WLERSAGLRVTMHQPVL-LCLLAFIWLMMK---
PRND_ponPyg -MKNHLS-WWLATVCMLLFSHLSAVQARGIKHRIKWNRKALPSTA-QITEA-PGAENRPGALSKQGRQLD-IDFGTEGNRYYEANYWQFPDGIHYDGCS---EANVTKEVFVTGCINATQAANQGEF--QKPDNKLHQQVLWRLVQELCSLKHCEFWLERGAGLRVTMHQPVL-LCLLAFIWLMVK---
PRND_nomLeu -MRKHLSWWWLATVCMLLFSHLSAVQARGIKHRIKWNRKALPSTA-QITEA-QVAENRPGGFIKQGRKLD-IDFGAEGNRYYEANYWQFPDGIHYDGCS---EANVTKEVFVTGCINATQAANQGEF--QKPDNKLHQRVLWRLVRELCSLKRCEFWLERGAGLRVTMHQPVL-LCLLAFIWLMVK---
PRND_papHam -MRKHLSWWWLATVCMLLFSHLSAVQTRGIKHRIKWNRKALPSTA-QITEA-QVAENRPGAFIKQGRKLD-IDFGAEGNRYYEANYWQFPDGIHYNGCS---EANVTKEAFVTGCINATQAANQGEF--QKPDNKLHQQVLWRLVQELCSLKHCEFWLERGAGLRVTMHQPVL-LCLLALIWLTVK---
PRND_macMul -MRQHLSRWWLATVCMLLLSHLSVVQARGIKHRIKWNRKALPSTA-QITEA-QVAENRPGAFIKQGRKLN-IDFGAEGNRYYEANYWQFPDGIHYDGCS---EANVTKEVFVTGCINATQAENQGEF--QKPDNKLHQRVLWRLVQELCSLKRCEFWLERGAGLRVTMHQPVL-LCLPAFVWLMV----
PRND_calJac -MRKHLSGWWLATVCMLLFSHLSAVQARGIKHRIKWNRKALPSAA-QITEA-QVAENRPGTFIKQGRKLD-INFGAEGNRYYEANYWQLPDGIHYDGCS---ETNVTKEVFVTGCINATQAANQAEF--QKPDNKLHQRVLWRLVQELCSLKRCEFWLERGAGFRVATHQPLL-LCLLAFIWLMAK---
PRND_tarSyr -MRKPLGGWRLAVVCVLLLGLLSAARARGIKHRFKWNRKASPSAA-RVTEA-RVAESRPGAFIRRGRKLD-IDFGAEGNKYYEANYWQFPDGIHYDGCS---KANVTKELLVAGCINATHAANAAERPREARDDPLHQRVLGRLVRELCSAKHCDFWPERAAAPGTARARP---GALLLVSRL------
PRND_micMur -MRKHLGGWRPAIVFILLFSHLAVVKARGIKHRIKWNRKPLPSTV-HITEA-LVAENRPGAFIKQGRKLD-VDFGEEGNRYYEANYWQFPDGIHYDGCS---DANVTKEVFVASCINATVAANQAEFAREKQD-KLHERILWRLTRELCSVKRCEFWLERGAGLRVAEDCPTI-LSLLLLLWLLLR---
PRND_tupBel --------------------------------------RALPSTA-HITEA-RVAENRPGAFIRQGRKLD-IDFGVEGNRYYEANYWQFPDGIHYDGCS---EVNVTKEMFITSCINATQAANQAEFSKEKQDNKLYQRVLWRLIKELCSIKHCDFWLERGAGLRVTMHLPVM-LCLLVFVWFVVK---
PRND_musMus -MKNRLGTWWVAILCMLLASHLSTVKARGIKHRFKWNRKVLPSSGGQITEA-RVAENRPGAFIKQGRKLD-IDFGAEGNRYYAANYWQFPDGIYYEGCS---EANVTKEMLVTSCVNATQAANQAEFSREKQDSKLHQRVLWRLIKEICSAKHCDFWLERGAALRVAVDQPAM-VCLLGFVWFIVK---
PRND_ratNor -MKNRVGTWGLAILCLLLASHLSTVKARGIKHRFKWNRKVLPSSG-QITEA-QVAENRPGAFIKQGRKLD-IDFGAEGNKYYAANYWQFPDGIYYEGCS---EANVTKEVLVTRCVNATQAANQAEFSREKQDSKLHQRVLWRLIKEICSTKHCDFWLERGAALRITVDQQAM-VCLLGFIWFIVK---
PRND_cavPor -MKTSLCVW--ALVCVLLQCHFSSVAARGIKHRIRWGRKPTPSPS-QVTEA-RVAMTRPGAFIKEGHKLN-IDFGAEGNRYYETNYWQFPDGIHYNGCS---DTNVTKEVLVTSCINATQAANQAEFSREKQDNKLHQRILWRLIKELCSA--------------------------------------
PRND_speTri -MKKHLGTWGLAVVCVLLSTHLSAIKARGIKHRIKWNRKSVPSTV-QITEA-QVAQNPPGAFIKQGRKLH-IDFGAEGNKYYEANYWQFPDGIYYDGCS---EANVTKEVFIAKCINATQAANQAEFSREKQDNKLHQRVLWRLIKELCSVKHCDFWLEGGAGFQLSVDQPVM-LCLLVFIWLMVK---
PRND_oryCun -MRKHLGAWGLAIVCVLLFSHLPPAKARGIKHRIKWNRKALPSTA-QITEA-HVAENRPGTFIRQGRKLN-IDFGPEGNKYYEANYWQFPDGVHYDGCS---EGNVTKEAFVAKCVNATQAANQGEFSREKQDNKLHQRVLWRLIKELCSIKHCDFWLERGAGVQVGGEQPLV-LGLLLCTWLLVK---
PRND_ochPri -MRKHLGGWGLAIVCVLLFSLLPTVKARGIKHRIKWNRKALPSTA-QITEA-RVAENRPGTFIRQGQKLS-IDFGPEGNKYYEANYWQFPDGIHYDGCS---EGNVTKEVFVAKCINATQAANQGEFSREKQ-NKLHQRVLWRLIKELCSIKHCDFWLERGAGFPVTVGQPMVVLCLLIFTWVLVK---
PRND_canfam -MRKHLGGCWLAIVCVLLLSQLSAVEARGIKHRIKWNRKALPGTS-QVTEA-RSAEIRPGAFIRQGRKLD-IDLGPEGNRYYEANYWQFPDGIHYNGCS---EANVTKEKFVTGCINATQVANQEELSREKQDNKLHQRVLWRLIRELCSVKRCDFWLERGAGPRVAGAQPVL-LCLLAFIWFIVK---
PRND_ailMel -MRKHLGGCWLAIVCVLLFSQLSAVKARGIKHRIKWNRKALPSTS-QVTEA-RTAEIRPGAFIRQGRKLD-IDLGAEGNRYYEANYWQFPDGIHYNGCS---EANVTKEKFVSGCINATRVANQEELSREKQDNRLYQRVLWRLVRELCSVKHCDFWLERGAGLRVGGDQALL-LCLLVFIWFIVK---
PRND_felCat -MRKHLGGCWLAIVCVLLFSQLSAVKARGIKHRIKWNRKTLPSIS-QVTEA-HTAEIRPGAFIRQGRKLD-IDLGAEGNRYYEANYWQFPDGIHYNGCS---EANVTKEKFVTSCINATQVANQEELSREKQDHKLYQRVLWRLNRELLWSAHGNFGRERG----------------------------
PRND_equCab -MRKHLGGCCLAIVCVLLFSQLPVVLSRGTKHRIKWNRKALPSTA-KVTEA-RVEEIRPGAFIRQGRKLN-INFGAEGNRYYEANYWQFPDEIHYNGCS---EANVTKEKFVISCINATQEANQEELSQEKQDDKLYQRILWRLISELCSVKHCDFGLESGTGLRVTMDQPVL-LCLLVFIWFIVK---
PRND_tapTer --------------------------------------KALPSTS-KVTEA-HTAEIRPGAFIRQGRKLN-IDFGAEGNRYYEANYWQFPDEIHYNGCS---EANVTKEKFVISCINATQAENQEELSREKQDDKLHQRILWRLIRELCSV--------------------------------------
PRND_sorAra -MRKHLG-WCLAIACVLLLCQVPAAQARGIKHRIKWGRKAPPSTP-QVTEA-RVAEIRPGAFIRQGRKLN-LNLGGEDKRYYDAYYWQFPDGIHYNGCS---SANVTREKFISGCINATRAANQEELSREKHD-KLYQRVLWRLVRELCSLKRCDFWSERGAGLQVTVDQLMM-FCLLAFTWFIMK---
PRND_eriEur -MRKHLGGWWLAIVCVLFFSQLSAVKARGIKHRFKWNRKALPSTN-HVTEA-QVSEIRPGAFIRQGTKLD-IDLGAEANRYYEANYWQFPDGIHYNGCS---EVNVTKAKFIASCINATHSANQEELSREKHD-KLYQRVLWRLVRELCSLKRCDFWSERGAGLQVTVDQLMM-FCLLAFTWFIMK---
PRND_cynSph --------------------------------------KTLPSPSSHVTES-QTAEMRPGAFVKQGRKLN-IDFGDEGNRYYEAHYWEFPDGIYYDACS---KANVTKEKFVTSCINATQAVNQEELSQEKQDKLLHQRILWQLIRELCSV--------------------------------------
PRND_bosTau -MRKHLGGCWLAIVCILLFSQLCSVKARGIKHRIKWNRKVLPSTS-QVTEA-RTAEIRPGAFIKQGRKLD-IDFGVEGNRYYEANYWQFPDGIHYNGCS---KANVTKEKFITSCINATQAANQEELSREKQDNKLYQRVLWQLIRELCSTKHCDFWLERGAGLRVTLDQPMM-LCLLVFIWFIVK---
PRND_oviAri -MRKHLGGCWLAIVCVLLFSQLSSVKARGIKHRIKWNRKVLPSTS-QVTEA-HTAEIRPGAFIKQGRKLD-INFGVEGNRYYEANYWQFPDGIHYNGCS---EANVTKEKFVTSCINATQVANQEELSREKQDNKLYQRVLWQLIRELCSIKHCDFWLERGAGLQVTLDQPMM-LCLLVFIWFIVK---
PRND_capHir -MRKHLGGCWLAIVCVLLFSQLSSVKARGIKHRIKWNRKVLPSTS-QVTEA-HTAEIRPGAFIKQGRKLD-INFGVEGNRYYEANYWQFPDGIHYNGCS---EANVTKEKFVTSCINATQVANQEELSREKQDNKLYQRVLWQLIRELCSIKHCDFWLERGAGLQVTLDQPMM-LCLLVFIWFIVK---
PRND_conTau -MRKHLGGCWLAIVCVLLFSHLSSVKARGIKHRIKWNRKVLPSTS-QVTEA-RTAEIRPGAFIKQGRKLD-IDFGVEGNRYYEANYWQFPDGIHYNGCS---EPNVTKENFITSCINATQVANQEEMSREKQDNKLYQRVLWQLIRELCSIKHCDFWLERGAGLQVTLDQPMM-LCLLVFIWFIVK---
PRND_girCam -MRKHLGRCWLAIVCVLLFSQLSSVKARGIKHRIKWNRKVLPSTS-QVTEA-HTAEIRPGAFIKQGRKLD-IDFGVEGNRYYEANYWQFPDGIHYNGCS---EANVTKEEFVTSCINATQAANQEELSREKQDNKLYQRILWRLIRELCSIKHCDFWLERGAGLQVTLDQPMM-LCLLVFIWFIVK---
PRND_ama -MRKHLGGCWLAIACILLFSQLSSVKARGIKHRIKWNRKVLPSTS-QVTEA-HTAEIRPGAFIKQGRKLD-IDFGVEGNRYYEANYWQFPDGIHYNGCS---EANVTKEKFVTSCINATQVANQEELSREKQDNKLYQRVLWQLIRELCSIKHCDFWLERGAGLRVTLDQPMM-LCLLVFIWFIVK---
PRND_turTru -MRKHLGGCWLAIVCVLLFSQLSSVKARGIKHRIKWNRKALPSTS-QVTEA-HTAEIRPGAFIRQGRKLD-INFGAEGNRYYEANYWQLPDGIHYNGCS---EANVTKEKFVTSCINATQAVNQEELSREKQDNSLYQRVLWQLIRELCSNKQCDFWLERGAGLRVTLDQPMM-LCLLVFIWFIVK---
PRND_pseCra -MRKHLGGCWLAIVCVLLFSQLSSVKARGIKHRIKWNRKALPSTS-QVTEA-HTAEIRPGAFIRQGRKLD-INFGAEGNRYYEANYWQFPDGIHYNGCS---EANVTKEKFVTSCINATQAVNQEELSREKQDNNLYQRVLWQLIRELCSNKQCDFWLERGAGLRVTLDQPMM-LCLLVFIWFIVK---
PRND_phyCat --------------------------------------KALPSTS-QVTEA-HTAEIVPGAFIKQGRKLD-IDFGAEGNRYYEANYWQFPDGIHYNGCS---EASVTKEKFVTSCINATQEVNQEELSLEKPDNKLYQRVLWRLIRELCSI--------------------------------------
PRND_susScr -MRKHLGGRRWAIVCILLFSQLSEVKARGIKHRIKWNRKALPSTS-QVTEA-HTAEMRPGAFIKQGRKLD-IDFGAEGNRYYEANYWRFPDGIHYNGCS---EVNVTKEKFVTSCINTTQAANQEE-SHEKPDNKLYQRVLWRLIRELCSIKHCDFWLERGAGLRVTMDQPMM-LCLLVFIWLIVK---
PRND_borAnc -MRKHLGAWWLAIVCILLFSHLSTVKARGIKHRIKWNRKALPSTA-QVTEA-HVAEIRPGAFIRQGRKLD-IDFGAEGNRYYEANYWQFPDGIHYNGCS---EANVTKEMFVTSCINATQAANQEEFSREKQDNKLHQRVLWRLIRELCSIKHCDFWLERGAGLRVTLDQPVM-LCLLVFIWFMVK---
PRND_dasNov -MRKHLGGWRLAIVCVLLSGHLSMVKARGIKHRIKWNRKAAPGAA-QVTEA-RVAEQRPGAFVRQGRRLD-IDFGAEGNRYYEANYWQLPDGILYDGCA---EANVTKEALVAGCVNATQLANQAELAHEGQDT-LHRRVLGRLIRELCALKRCKFWPDRAAGPRLVRGAPVF-GGLLLLIWLLVR---
PRND_choHof -MRKHLGGWWLAAVFVLLVCHLSVAKARGIKHRIKWNRKALPSAA-QVTEA-RIAESRPGAFIKQGRKLD-IDFGAEGNKYYEANYWQFPDGIHYDGCS---EANVTKEVFVTGCINATQAANQAEFSREKHD-KLHQRVLWRLIRELCSVKHCDFWLERGGGLRLTVDQPVV-SACWFHLAYRE----
PRND_losAfr -MRKHLGAWWLAIAFVLLLSHLSMVTARGIKHRIKWNRKALPNTG-HVTAA-QVTETRPGAFIRHGRKLD-IDFGAEGNRYYEANYWQFPDGIHYDGCS---EANVTKEMFVTSCINTTQAANQEEFSR-KQDNKVYQRILWRLIRELCSVKHCDFWLDRGGGLRVSLDQPVM-LCLLVFIWFMVK---
PRND_eleMax --------------------------------------KALPNTG-HVTAA-QVTETRPGAFIRHGRKLD-IDFGAEGNRYYEANYWQFPDGIHYDGCS---EANVTKEMFVTSCINTTQAANQEEFSREKQDNKVYQRILWRLIRELCSV--------------------------------------
PRND_proCap --------------------------------------KPVTNPA-HMTAA-QVSDRRLGTFIRHGRKLD-IDFGAEGNRYYEANYWLFPDGIHYDGCS---DTNVTKELFVTNCINTTQAVNQEEFSREKQDNKLYQRILWRLIRELCSLLGLDFWPERVGGLFFSLDQPVM-LCLLVFIWFMLK---
PRND_echTel -MRKHLGAGWLALACVLLFGHLSVVPARGIKHRIKWNRKTLPSTA-HVTEA-RVSVTRPGAFVRHGQKLD-VDLGADGNRYYEAHYWQFPDGIHYDGCS---DANVTREMLVTRCINATQAANQAEFSRDQQDSRLHQRVLWRLIRELCSAKRCDFWLEGAGASRAGLDQPPVLLGLLVLLGLTVK---
PRND_triMan --------------------------------------KPLPNTP-HVTAA-QVSDARPGAFIRHGRKLD-IDFGAEGNRYYEANYWQFPDGIHFDGCS---EANVTKEMFVTSCINTTQAANQEEFSREKQDNKLYQRILWRLIRELCSV--------------------------------------
PRND_monDom -MRRHLGICWIAIFFALLFSDLSLVKAKTTRQRNKSNRKGLQTNRTNPTTV-QPSEKLQGTFIRNGRKLV-IDFGEEGNSYYATHYSLFPDEIHYAGCA---ESNVTKEVFISNCVNATRVINKLEPLEEQNISDIYSRILEQLIKELCALNYCEFRTGKGTGLRLSLDQYVM-VYLVILTCLIVK---
PRND_macEug -MRRHLGTWWTAIFFALLFSDLSLVKAKGTRQRNKSNRKSLQTNRVNPTTA-QPSEILQGAFIRQGRKLS-IDFGEEGNSYYETHYQLFPDEIHYVGCT---ESNVTKDIFISNCMNATHAVNNLETLEEKNASDIHSRVLEQLIKELCALKYCELETETGAGLKLSLDQSVM-VYLVILTCLIVK---
PRND_sacHar -MRTPLETWWIAIFFTLLFSDLSLVKAKGIRQRNKSNRKSLQTNRANPTRE-QPSKILQGTFIRKGRKLS-INFGEEGNSYYEAHYKLFPDEIHYVGCA---ESSVTKDVFISNCVNVTHTANKLEPPEERNSSAIYSRVLEQLIKELCALKYCEFGMQIGAGFRLSLDQSMM-VYLMILAFFIVK---
PRND_ornana MRRRSGGARWLLVFLVLLSGDLSSLQARGPRPRNKAGRKPPPSNAGPDSPAPRPPAGARGTFIRRGGRLS-VDFGPEGNGYYQANYPLLPDAIVYPDCP---TANGTREAFFGDCVNATHEANRGELTAGGNASDVHVRVLLRLVEELCALRDCGPALPTGPAPRPGPPGPPAALALLTLVLLGAQ---
PRND_chrPic MGRTRMVTCWMAMLLLVLCSNITLCR-RGSSSKKASQNKSPPPVNKPSPPTKKEPVKIPGTLCYTGQLLD-IKLEPEEAKYYTDNFKKFPDCIYYPRCFQSLEPNATKDTLVSECFNVTVSENKLDLPEGKNATSVYSRVMWQVISHLCAMEFCCQPCSLALSIHGSFGWLAMLC-LGSFISLTMQ---
PRND_anocar ---MMQRPLVVAILLTALWSEVCLCR-RVSGSANRRNKKTSTTTSAPKLQSSTTATTFQGNLCRGGQMIDNMDLEPNDKVYYKANLKIFPDGLYYPNCSLLLQPNTTKEELVGECVNFTIASNKLNLSKGKDLSNTKERVMWVLIHHLCANESCGQPCPLLQNSGNLHYIGQVLTVFVGLIGCSFLSAK
PRND_pytMol ----MWKNLLVVVLLAVLCSNVGLCR-RRSGPTRKWPEKTVS-------KSSTKSSTTLGNLCYGSEMIDNMILEANDTTFYKDNAKEFPNGLFYPNCSLPLQPKVSRETRTEECVDFIIASSNLTLPNGKNASDPSKRVMWLVINHLCASETCSLPCPGSSNVSSAHYFLPIL--MTALVGYSILATR
PRND_xenTro MGRQNLFSCLILLLLILYCS-LSSPR-RAASSKKI--SKTTDLSRGAKRRPKVTNSPALGDLSFRGRALNVNFNLTEESELYTANLYSFPDGLYYPRPAH-LSGAGGTDEFISGCLNTTIERNKVWISEDDEEGDIYMSVATQVLQFLCMENYVKPTNGAVTCTGGLWVFIGVMHFFFLFRKGD-----
* * * * ** *** * * * * * * * **