LOXHD1 SNPs: Difference between revisions
Tomemerald (talk | contribs) |
Tomemerald (talk | contribs) mNo edit summary |
||
| Line 2: | Line 2: | ||
=== Introduction === | === Introduction === | ||
=== 3D Structure === | === 3D Structure === | ||
Revision as of 16:16, 6 November 2009
LOXHD1
Introduction
3D Structure
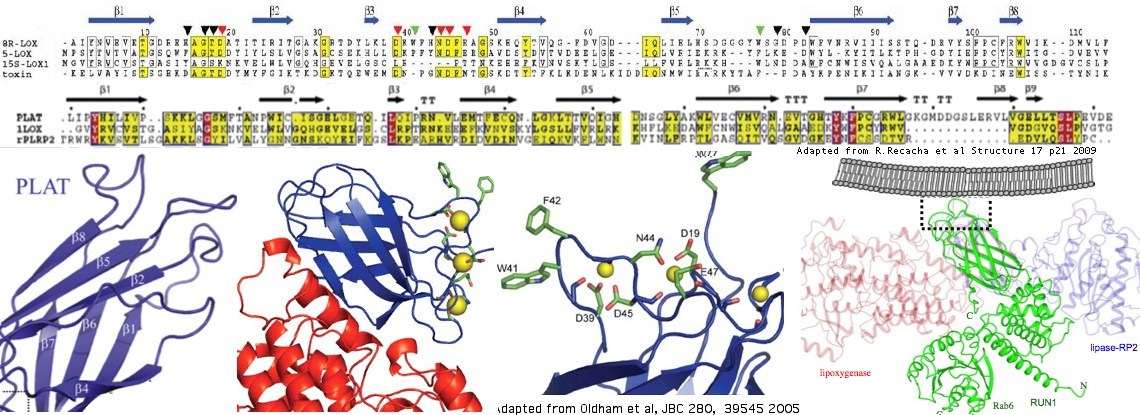
The structure of three proteins containing single PLAT domains have been determined. The best blastp of LOXHD1 to these is a coral lipoxygenase called allene oxide synthase 8r-lipoxygenase with PDB accession 3DY5. Lower quality but still useful matches occur with RAB6 (accession CWZ) and rabbit reticulocyte lipoxygenase (accession). The basic structure consists of a sandwich of two 2-stranded beta sheets (beta barrel) with certain conserved hydrophobic residues reaching out to the membrane. The lipoxygenases are calcium-binding via extended acid residues but these are not conserved in LOXHD1.
The PLAT domain is variously denoted as LH2 or C2 depending on the publication and domain tool (eg Pfam and SMART). The secondary structure is so simple that convergent evolution (independent origins) must be considered, as with TIM domains. Here however significant blastp matches at the primary sequence level cannot have originated independently given the vast combinatorial space available to amino acid sequences. Further, all PLAT/LH2/C2 domains share characteristic conserved residues. Thus these domains are all homologous (descended from a single source) and multiple names are inappropriate. PLAT is used here because it is an acronym of all protein classes containing the domain.
Below PLAT domains from all human proteins are aligned with the 16 domains of human LOXHD1. The coral lipoxygenase is included because it provides the template for beta strand assignment and extended residues. The lower alignment is just with human LOXHD1 domains and uses an optimized seventh PLAT domain (lacking the KE expansion).
Individual PLAT domains are quite conserved. That is illustrated by a difference alignment of the fifth PLAT domain in 40 vertebrates (human to lamprey) below. Note columns where human sequence represents a unique change -- such as H->R in column 5 -- may be minor alleles or disease variants rather than the predominate residue in the overall human population.
01.hg18_6 MARYHVTVCTGELEGAGTDANVYLCLFGDVGDTGERLLYNCRNNTDLFEKGNADEFTIESVTMRNVRRVRIRHDGKGSGSGWYLDRVLVREEGQPESDNVEFPCLRWLDKDKDDGQLVRELLPSDSSATLK 02.panTro ....R.............................................................................................................................. 03.gorGor S..R...........................................................................--------------------------..................G...... 04.ponAbe ....R..............T.....................................................................................-....................N.... 05.rheMac ....R.........................................................................................................................N.... 06.calJac ....R...........................................................K.............C...............................................N.... 07.micMur ....R...........................................................M.............................................................N.... 08.otoGar ....R. ............K.................-..........H.P.V.......T....................N.... 09.tupBel ...R...........................................................K............A................................................N.... 10.mm9_6_ ....R...........................................................K.....V.......................................................N.... 11.rn4_6_ ....R...........................................................K.....V........................................Q................... 12.dipOrd ....R.....................Y..................E..................K.....V.......S.S.............................................N.... 13.cavPor ....R.......F.............Y...................................L.K.............................................................N.... 14.speTri .V..R.....................Y......-.....-------------............K............P......E...L.....................................N.... 15.oryCun ..........S.............K..........................................................TG.N.... 16.ochPri ....R...........................................................K.............S....V..........................................N.... 17.vicPac ...R.......V.............................I.....................K.................F.E.........................................N.... 18.turTru ....R...........................................V...............K.....V...........F.......................---------...........N.... 19.bosTau ....R.....................Y.....................................K.................F.E....................-....................N.... 20.equCab ....R...........................................................K............A....F.E.........................................N.... 21.felCat ....R...........................................................K............G....F.E.........................................N.... 22.canFam ....R...........................................................K............G....F.E.........................................N.... 23.myoLuc ...HR......D.R......S.....Y............T.K..DEM.Q...............K........T.V.G....F..................Y........................N.... 24.pteVam ...R.........................................M.................K..........R.G....F.E.........................................N.... 25.eriEur ....R.....................Y.....................................K............G....F.A..............M..........................N.... 26.loxAfr ..Y.......-..M.....................A..L.K............G......E.........................................N.... 27.proCap ....R.....................Y..A.......M...........R.........A..L.K............A......E..............M...---...................NN.... 28.echTel ....R.....................Y..........MF.....V..............A..L.K......M.....A......-...LKK.........K..G.G....................N...R 29.choHof .V..R.....................Y.........W.........M....S...Y...A..L.K.K.I.VG.....GN...F.E..............M....N...AR.Q.....I......G.N.... 30.monDom ..Q.R..L...DI.....N.QAFV.....A......M.......MEICQ..........A..V.K......G.....K....F.VK..................N..F...Q..............NSR.. 31.ornAna IVK.R...V..D.N......R.FI..I.........I.........T.........FV.A..LKQ......G.....GS.....AK.I.........EA.....Y.....NE....I....V.AGE.PL.. 32.galGal VIK.R......MVS.S......FV..I..Q....D.V..K.I..VNK.........F..A..LKQ......G.....GS.....AK.I.........EA.....Y.....NE....I....V.AGE.PL.. 33.anoCar IK.R...H..NVS.S....H.FA..I..Q....D.V.Q.SV.TVNK....S....I..A.NLKQ......G.....GS.....GK.I...D.....EAQ....Y.....NE....II...V.AGR.SL.E 34.xenTro ...F.IE.K..MNK.......D.I..A..LKQ.K....G.....G.C...M.K.V...D.K..TEAI....N..F.RNE...HII...V.AGD.QN.R 35.tetNig LK.RI.I...NVG.G....S.F.NII..L......SMITSK..VNK.....H...L....CLGR.S...VG...R.G.C..F..K.T.........SAI....F....HNE....I....V.AGDGRLF 36.fr2_6_ LIK.R..I...NVS.G....S.F.NVI..L.......MIMSK..VNK.....H...L....SLGQ.....VG...R.G.C..F..K.M.........SAI....F....HNE....I....V.A 37.gasAcu LIK.R..I...NVS.S....S.F.NVI..L.......MFMSK..VNK.....H...L....SLGQ.....VG...R.G.C..F..K.M...D.....LAT....F....RNE....I....V.AGDGRLF 38.oryLat IK.R..I...TVS.S....S.F.NVI..L.......M.MSK..VNK.....H...L....WLGQ.....VG...R.G.C..F..K.M.........AAI....N....QNE....II...V.AGEG 39.danRer VK.R...Y..DVS.S....H.F...I..L....D.S.I..K..INK..R......I..A.SIGP......G...R.G.C.....K.TI.....A..QA.....S..F.RNE....I....Q.FADGRLY 40.petMar VK.L..IH..KHG.G......FV..Y.EQ....D.M..TS...VNK..R..V...V..C..LKR.T....G...R.GS...F..KIM.K-D.RRQCEE.....N....D.E....I....VAGGCQMLK Consensus ...yrvt..tg...g.gtda.v..cl.gd.gdtger.ly.crnn...fekgna...t..s.t$rk.r...!r...k.ggs..y$d.!lvre#.qpes.nv...c.r.l.k#k....l!...l.sgsnatlk
Reference sequences (intronated)
The sequences below are broken into individual exons with reading phase (codon overhang) indicated by 012. Alternating colors show odd and even numbered PLAT domains. The 7th domain (which contains the charged KE expansion loop in mammals) is shown in red.
>LOXHD1_homSap_full 41 exons|2273 aa|chr18|span 179,559 bp|no signal peptide|no conserved cysteines|16 PLAT domains|EIW --> WNW mouse deafness
0 MMPQKKRRRKKDIDFLALYEAELLNYASEDDEGELEHEYYKAR 1
2 VYEVVTATGDVRGAGTDANVFITLFGENGLSPKLQLTSK 2
1 SKSAFEKGNVDVFRVRTNNVGLIYKVR 2
1 IEHDNTGLNASWYLDHVIVTDMKRPHLRYYFNCNNWLSKVEGDRQWCRDLLASFNPMDMPR 1
2 GNKYEVKVYTGDVIGAGTDADVFINIFGEYGDT 1
2 GERRLENEKDNFEKGAEDRFILDAPDLGQLMKINVGHNNKGGSAGWFLSQ 0
0 IVIEDIGNKRKYDFPLNRWLALDEDDGKIQRDILVGGAETT 1
2 AITYIVTVFTGDVRGAGTKSKIYLVMYGARGNKNSGKIFLEGGVFDRGRTDIFHIELAVLLSPLSRVSVGHGNVGVNRGWFCEK 0
0 VVILCPFTGIQQTFPCSNWLDEKKADGLIERQLYEMVSLRKKRLK 1
2 KFPWSLWVWTTDLKKAGTNSPIFIQIYGQKGRTDEILLNPNNKWFKPGIIEKFR 0
0 IELPDLGRFYKIRVWHDKRSSGSGWHLER 0
0 MTLMNTLNKDKYNFNCNRWLDANEDDNEIVREMTAEGPTVRRIMG 1
2 MARYHVTVCTGELEGAGTDANVYLCLFGDVGDTGERLLYNCRNNTDLFEKGN 0
0 ADEFTIESVTMRNVRRVRIRHDGKGSGSGWYLDRVLVREEGQPESDNVEFPCLR 2
1 WLDKDKDDGQLVRELLPSDSSATLK 1
2 NFRYHISLKTGDVSGASTDSRVYIKLYGDKSDTIKQVLLVSDNNLKDYFERGRVDEFTLETLNIGN 0
0 INRLVIGHDSTGMHASWFLGSVQIRVPRQGKQYTFPANRWLDKNQADGRLEVELYPSEVVEIQK 1
2 LVHYEVEIWTGDVGGAGTSARVYMQIYGEKGKTEVLFLSSRSKVFERASKDTFQ 0
0 LEAADVGEVYKLRLGHTGEGFGPSWFVDTVWLRHLVVREVDLTPEEEARKKKEKDKLRQLLKKERLKAKLQRKKKKRKGSDEEDEGEEEESSSSEESSSEEEEMEEEEEEEEFGPGMQEVIEQHKFEAHRWLARGKEDNELVVELVPAGKPGPE 1
2 RNTYEVQVVTGNVPKAGTDANVYLTIYGEEYGDTGERPLKKSDKSNKFEQGQ 0
0 TDTFTIYAIDLGALTKIRIRHDNTGNRAGWFLDRIDITDMNNEIT 2
1 YYFPCQRWLAVEEDDGQLSRELLPVDESYVLPQSEEGRGGGDNNPLDNLALEQK 1
2 DKSTTFSVTIKTGVKKNAGTDANVFITLFGTQDDT 1
2 GMTLLKSSKTNSDKFERDSIEIFTVETLDLGDLWKVRLGHDNT 1
2 GKAPGWFVDWVEVDAPSLGKCMTFPCGRWLAKNEDDGSIIRDLFHAELQTRLYTP 1
2 FVPYEITLYTSDVFAAGTDANIFIIIYGCDAVCTQQKYLCTNKREQKQFFERKSASRFIVE 0
0 LEDVGEIIEKIRIGHNNTGMNPGWHCSHVDIRRLLPDKD 0
0 GAETLTFPCDRWLATSEDDKKTIRELVPYDIFTEKYMKDGSLRQVYKEVEEPLD 1
2 IVLYSVQIFTGNIPGAGTDAKVYITIYGDLGDTGERYLGKSENRTNKFERGT 0
0 ADTFIIEAADLGVIYKIKLRHDNSKWCADWYVEKVEIWNDTNEDEFLFLCGRWLSLKKEDGRLERLFYEK 0
0 EYTGDRSSNCSSPADFWEIALSSKMADVDISTVTGPMADYVQEGP 1
2 IIPYYVSVTTGKHKDAATDSRAFIFLIGEDDERSKRIWLDYPRGKRGFSRGSVEEFYVAGLDVGIIKKIE 0
0 LGHDGASPESCWLVEELCLAVPTQGTKYMLNCNCWLAKDRGDGITSRVFDLLDAMVVNIGVK 0
0 VLYEMTVWTGDVVGGGTDSNIFMTLYGINGSTEEMQLDKKKAR 2
1 FEREQNDTFIMEILDIAPFTKMRIRIDGLGSRPEWFLER 0
0 ILLKNMNTGDLTMFYYGDWLSQRKGKKTLVCEMCAVIDEEEMMEWTSYTVAVKTSDIL 1
2 GAGTDANVFIIIFGENGDSGTLALKQSANWNKFERNNTDTFNFPDMLSLGHLCKLRVWHDNK 1
2 GIFPGWHLSYVDVKDNSRDETFHFQCDCWLSKSEGDGQTVRDFACANNKICDELEET 1
2 TYEIVIETGNGGETRENVWLILEGRKNRSKEFLMENSSRQRAFRK 2
1 GTTDTFEFDSIYLGDIASLCVGHLAREDRFIPKRELAWHVKTITITEMEYGNV 2
1 YFFNCDCLIPLKRKRKYFKVFEVTKTTESFASKVQSLVPVKYEVIVTTGYEPGAGTDANVFVTIFGANGDTGKRELKQKMRNLFERGSTDRFFLETLELGELRKVRLEHDSSGYCSGWLVEKVEVTNTSTGVATIFNCGRWLDKKRGDGLTWRDLFPSV* 0
>Gallus gallus XM_425221 14 LH2 domains MEQILLKSLLRHVENKDEVIDVYEVVTVTGDVRGAGTDANVFVTLFGEFGITPKTHLTSKSSTAFERSKTDVFRVKTNNVGQIKKIRIEHDNTGLNAGWF LDRVIVTDMNRPHLRFYFPCNNWLSKEDGDGLYVRDLIGSLNPMDVPKINKYVVRVFTGEVSGSGTDADVFINIFGEKGDTGVRKLDNDKDNFEKGAEDK FTLDAPNLGRLRKINIGHNNKGGSAGWFLAKVIIEDIGNKCVYQFPVGRWFALDEDDGKIQRDILVGGTEATGIVYNVAVVTGDIRGAGTNSKIHVILHG SKGLKNSGTIFLEGGEFERARTDLFNVEIASLLSPLSRVTIGHDNCGVSSGWYCEKVVVYCPFTGIEQTFPCGKWLDEDEGDGLIERELYEMVSLRQRRL KKNPWSLWIWTSDIKNAGTDATIFFQIYGDKGKSDEMKLDNNSDNFEAGQTDKFMIELPDLGTFYKLRIWHEKRNPFAGWHLDKVTLLKTLTKDKYSFNC GRWLDINEDDNEIVRELPAEGSLVTEVMPVIKYRVTVCTGMVSGSGTDANVFVCLIGDQGDTGDRVLYKCINNVNKFEKGNADEFFVEAVTLKQVRRVRI GHDGKGGSSGWYLAKVIVREEGQPESEAVEFPCYRWLDKNEDDGQIVRELVPAGESPLLKNVSYHISVKTGDIPGASSDSKVFIKLYGEKADTSKETLLV SDNDLGNYFERGRVDEFTIDTMDIGKINRILIGHDNVGLRSGWFLASVQITVPVQGRQYMFPCNRWLDKDEADGRVEVEVYPSEILPIEKLINYEVSVVT GDVRAAGTNAKVFMQIYGETGKTELIILENRSNNFERGATDIFEVEAADVGKIYKIRIGHDGKGIGDGWFLESVTLKRLATKMDETDKKKKKKKKKSEEE EEEEETKVEEVMDVYTFVAHRWLAKDEGDKELVVELVPDGESELEENTYEVHVLTGSVWGSGTDANVFLSIYGIERGDTGERQLKRSNNLNKFEKGQVDV FTIKAIDLGELKKLRIRHDNSGSSPSWFLERVEIVDLKESTTYYFPCQRWLAVEEDDGQIVRELVPVDEAFVKKDSENDGQSLATLGLEQKAKSTTYIVK VKTGDKKNAGTDANVFITLYGSKDDTGIVSLKASKLNKNKFERGKIDEFTVESVDIGDLKKIKIGHDNAGNSNGWFLEWVEIDAPSLGQCLKFPCGRWLD KSEDDGAIERFIFPAELQTTEYIPFVPYEITVYTSDIFGAGTDADVFIVLYGSDGICTQQKSLCLNKREQRMYFERNSVNQFIVELEDVGDIIEKIRIGH NGGGLNSGWHLDRVAIRRLLPNGKGSETITFPCERWLAKSEDDGEIIRELVPSDIFTEKLMKDGTLKQIEEEVEDPLEVHTYKISVFTGDIYGAGTDANV FLNIYGDLGDTGERKLSKSETNFNKFERGQSYDGERRSLSESSVSLSSLDSSGNPKEKKSRLVKSAEEGLLIPYHITVTTGTEYDSSTDSRVFIIIMGPQ KVRTERLWLDLPEGKDEFADGSVEKFSVWGLDVGEIKKVEILYEVTVVTGDIESGGTDAGIFMTVFGSNGNTEEMQLDKNGDRFERGQEDSFIMEIADIA PLRKMRIRTDAKGTRPDWFLERIVMRNLTNQEVATFTYGDWLSKVKNAKGSLVCEMPAMVNDEQMMEDTTYTIQVKTSDIGGAGTDANVSLILFGENGDS GTLALKESNKSNKFERNQMDEFNFPNMLSLGDLCKVRIWHDNKAYEIVTVTSNREDAETKENIWIILEGKLGRSKEFLMENSSKKRRFERGSTDTFQFSS KNLGDIAAICVGHCPKDGKKSSAKADVYWHVKEIIITEMELCNKYFFRCNGKIPLRYKRRDYKVFECAKVIESFASKARSLVPVKYETIVVTGFEKGAGT DANVFITIFGLNGDSGKRALKQKFRNLFERGKTNRFYLETLDMGELKKVRIEHDNSGLAPGWLVERVEITNSATGVTTIFPCGKWLDENRGDGLTWRELFPRY* >Schistosoma mansoni (roundworm) XP_002576380 2301aa Bilatera|Platyhelminthes blast 0 MNLEDSTNDDYLKNSKSRFSFHRKEEKEHKGIICCGHNTCHLDRYNLISSVKPFTARPYTLYKVVLTTADVPGAGTSAQ 1 2 IYITLKGEWGSSTRQKLRKEKVPTRNLRFYFYPGSTNTFSVVSPDLGGLHSVFIE 0 0 HDSLRKSDSWLLESVQVFHPLTKKRYMFMCNHWFSLYKEDGLIARELFGIRSAKT 1 2 KYSIVTVTGDQEGSGTNSKVYITIYGRTGITPRIELSQENKSTGKDILCAPFGRGTSTKFIVKAPNVGAITNIRIKQDESGNEPHWFLERVVVTDMSYPQWTYYFHLSCWLSSKYGDGKSCRLVRGYREPTGTGV 1 2 ETEYKLTFYTSDQKDAGTTGEVYVKLYGEKDSSREIWVNSINQKNRRQPTSYHFTRGSTVEVYLPPCPQLGEITKLKVGHNRAGSSPSWFLNK 0 0 VIVDDLRMNRVFEFPCYAWITKPSETIIVCQKSIKKEEKEIIWQ 1 2 KIPLEIRLYTGDVPNAGTTAKVYL 12 HQKTTSENKEDVYTTPYIWLEDGNYDRNGVTLFSIDLPVPKFISPLSQLVIGHDNTGHSPSWFFDK 0 0 VQIYCPLNGVEQTFLYRKWLTSSKPGMKVEQILCEEKSLRKCTDK 1 2 KIPWEISVKTSPIANSYVTAAVSIIIFGSKDKTMKIQLDRSNLEVSSEWTKVSGNEVVEMFRPNVESKFRIYIKDIGIPCKLRIQHDNKGSNPNWHLQE 0 0 IVLTNLRTHEQYEFYCNRWLSTREDDGSILREIPAKGPGITNPLP 1 2 LYHYIIQIFTGNKPNSGTNANIFINIFGEKGDCGERWLGRSVNRNSELFQQNQ 00 MDEFVIEAVQLGSINKICIGHEERSPGYGWYLAKIVLTIKENPKYKLTFECYR 21 WFDVGEDDGQIVRELFAHSSLS 1 2 AIAYNVTVLTGTCRNAGTVANVFVHLYGLQGESKDMQLKHKETEITKFEAGKSEEFILACGKLGE 0 0 IKKIKIWHDSIAPHSGWFLDEVVVIDYFLGRRYVFHVNRWLAKNEADGLISLDLQPTSIINIER 1 2 KSSYEVIVKTGGRKYAGTDSHVYITMFGINSESKEYHLANSKTHNNKFEQNHEDLFN 0 0 LNAVGLGSLKKIRVRHTNTGIAPGWFLDYILIRESIEQKKMEYFFPCYQWLSATRLDGLVIRELSVASENIFERWKSGHDISDDSRLILE 1 2 DQSTIYHVRIHTGSQSNISSDANVQIQLYGDKDFTGNIRLYKAYYGKEDILVNKFQAGQIANFIVKAINIGNIKKIR 2 1 IEHDTAVGSARWFVERVEVEARKLGLLWKFECNRFITPQQPELILYPEPKDISFVKP 1 2 MTNYQIKTFTSDISGAETTSSVYIQLYGNDSLPSSIRRLHQNNDSEQRFQRNKIDTFYVELEELQEPFSKLRIWHNDKGSSTDWHLNKVEIRKIKTNQY 0 0 VFVTYIFPCNKWLSRNMDQAALERELIPSHMLQDQNGVLQFERQLSPKWLMNTYEVRITTGDKAYAGTDASVYITLFGENGDSGERKLTKSLTHRNKFERGQ 0 0 TDVFQLEIVDLGKINKVRIRHDNSGVNPSWYLSTIEVFNISKSQSVHGLKDQPNETHHLVKYIFNCEAWLSTEHDEKVLDRFFSAEIIKPQDLPGIVPKTSKDIVPHKLIQDLSQGKKALE 1 2 TIPYLITVITGSDRKAGTPGPVWISCVDKDKMNSEKFILCDCYNRTMLKRGTTRYFRFAGVKLNDLTEIQ 0 0 VGNDAPESPSMGWYIQSLYLSFPTIGKMYLFDCKEWLSTNRGNKKRTCNLKISNTNIVKFKP 1 2 KITYHLKITTANVKRAGTDCSINLQIFGTNGVTNCYILEKTSNRFAQGITDNISLEMEDVGKLLKMRIGHDNQ 0 0 GKNKHWNLSCVEVTVANTNQLYRFVYDDWLSLTYGKRKSLWADLPAMIGDTVQLK 1 2 ETCLDIFVKTGNMPASSTDANVYVQLFGEYGDSGEILLKQTVSNQKPFQNNS 0 0 IDHFKIPSILKLGNLARCRIWHDNKGSSPNWYCEWLEVKEVLIPGEKNLACNWKFAFNKWLSVSDDNKQLLRDAPCSEVYMNDSKGHRTIDQESIETLLTTADSMNISDLKDNPEGKL 1 2 VYEVVIETGNLKDSGTTCDAWIILEGKHGRSPKLELVNQVGNPILQINQMNTFQ 2 1 LPSFPLGDLETIRLGIQERNINKQTNPNDIQSQKWFCEKVSIKDPVSKRTYIFTIHQWLSVTPTSNLKKDVLVNYSKIIEDPYKKAFNELK 1 2 NKQTVMYKVSIYTGTKGCANTDANIFITMFSTTPGLNSGRIALKRENNNLFDRKQLDEFYVESIDL 1 2 ESIQRIIIEHDNTGVSPDWYLDKVLITNQTNNQIHLFQCYQWISKKKGDCRLWKELLVSN* 0 >Trichoplax adhaerens (trichoplax) Metazoa XM_002107971 17 PLAT domains blast 0 MSASLAMQELMSDPSGHFAPKPPPGRSKINRKRPQT 1 2 APGRQITTASKAPKLWRSSTYNEPNLKRRKFIVTLNHKKDISSISRNAPYANRAYTTAMATAARTYSNALNKGQQKEIIPIYNPLCDPHLNDYYARKFGLLNSRD 0 0 ESRKNRKQ 1 2 AGVAYQFGVKTGDKKGSDTDA 2 1 VYIQVIGTKDKIPKKRLFKKQETEKTERGNLFKFDKSTVEKFAVQHRDIGDPVKLIVE 0 0 HDGNEKRHGWFLEEITLTNIQSKKSWLFPCHKWLSKYEGDRKLCYELKPLAKAGKA 1 2 VYEVSVLTGDKRGAGTDANVSVTLFGKHTSSPKIQLLKS 2 1 SKHKNPFERNNTDEFKIRTRDVGKLSKIRIEHDNAGFGPGWFLDK 0 0 VIICNLEKPNVKYYCPCNQWLAKDVGDKSISRDLTAYTDPNAAPS 1 2 AYVYIVHTFTGNKRGAGTDANVYAVIFGDSGDTGEKRLDNSKNNFEKSR 2 1 KDTFKLSCSCVGKLERLRIRHDNTGLFAGWYLDKV 1 2 VVEDPQEQQSYTFYCRRWLSKTEDDGEICRDLIVSASGDDGDDSVAPK 1 2 GYPYHIHVTTSDVKNAGTDAEVYVVMHGEGKKSKELNSGKLVLANSEKKKNTFERAMTDIFHMECAEMLSPLTKLTVGHDNKGLAAGWHLDR 0 0 IVIDCPTTGIEQTFLCQQWLDRKAGDGLTERELVEAFDMRKTRRP 1 2 KQLWFAWIWTSDIRGAGTDANVSMQIYGDKGKSQEIKLGNNTDNFEQATLDKFK 0 0 LEIDQVGVPYKLRIGHDNSNAFPGWHLDKVKLENMNDKEQYLFNCNRWLSRSEEDNEIIRELPASGPNCPNYP 1 2 IVIYEVSVHTGNKMGGGTDANVFIKIYGELGDSGYRPLKSSKSHNNKFERNQVDVFHIEAVTLKALKKIKIGHDGNNP 1 2 GAGWFLDKVVIKELNGEASNEFPCNR 2 1 WLSKSEDDGQIVRELFLKSDTPLLK 1 2 TTSYHISVKTGDVRNAGTDANVFIQIFGAKDDTGRVRLKQSLNTSNKFERNRIDKFIIEAAQIGK 0 0 IEKIIIGHDGKGLGSGWFLDYIELDVPSVGRLYRFSCHQWFDSTEGDRKVERELYPSECIKSAA 1 2 KIPYQISVHTGDIRHAGTDSNVFAVIYGENGKTEELKLRNKSDNFERGQVDVFK 0 0 VECEDVGKLRKLRIGHDSAGMGSAWFLDK 0 0 VYVRRLPPKSGKKSKETDEREEETAKKDADEPEKLDENNYLFVANRWLSKEEGDRQTVIEISPVGVDGALA 1 2 EMTYTIRVITGNKFGCGTNANVFINMYGEEGDSGERQLKKSETHTDKFERNQ 0 0 EDVFKISCLSLGELKKIKIRHDNSGFRPAWFLDKVIIEVGESKYQFMCDRWLAKDEDDGQISRELLPQSDEQSRAEAIGASKDLQKK 1 2 VASTTYNVSVTTGDIKGAGTDANVHIVLYGEKDDTGLIHLKNSTTHSNKFERNQEDRFVVEAIDIGELKKIK 2 1 IGHDNKGGMAGWFLNKVEIDIPSLGRRLLFPCGRWIDKGKDDGALERELYPLNEAEETYRP 1 2 HIPYEVTVYTTDKRGASTGANVYVVIYGEENQTEQASLEPDKKRRKQYFKNGAIDKFVLE 0 0 LDDVGEEITKLRIGHDGKGWGAGWHLDKVEICRLLDGGKASKKFTFQCNRWLASDEDDGAIVRELVPSEIVEKSSKDGGQVKTKVTKPTDGLK 1 2 VKPYTIHVFTGDVDGAGTNANVFLTIFGESGDSGERKLAKSDTHYDKFERNQ 0 0 EDIFHIEAADLGRLFKVKIRHDNTGSLFSPAWFLNRIEIVDDENEETTAFPCERWLAKKKDDGKIDRTLFVKGWEGDTSSVATTRSK 1 2 ASFRSNSTDLGEMVSASGRRASSRKGPVMAPIDEK 1 2 CVPYNIKVTVGEESSKNFQETLHLELFGQIEEEKSGPIELSPEKKSDKTFYPGKITTFYVSAAEVNIIEKIQVS 1 2 HNSYMPDSGIYLKEIEVDVPTIGNKYIFPCNRWLAKDKDDSKTSRIFTAANAQVTSYTA 1 2 GTDSNIFVILFGTKGRTPEISLEKNEDRFERAKVDIIP 0 0 LELDDVGTIKKIRIGHDGKGSRTDWYLEK 0 0 ASIQRMDTLDMYMFRANQWFSKKIDDKKLVREIPAETSKEGATTIK 1 2 KINYVLSTHTSDKRGSGTDANVFVIIFGENGDSGEIALKKSETNWNKFEKGQTDVFLINDRLSLGRLQKLRIWHDNA 1 2 GFGASWHLASVDIVDESTGVKYTFPCDKWLSKSNGDKLILRELPCAETAGNTAASKATKQESKSGKA 1 2 EYEIAFTTGTEKRAGTNQDVAIVLKGKSEKSREFLIENNEDKKYFSKGKTNKFTYTCKPLGDITKAIVSHRESAIGEEPDSKNSSWYLKVVTVVHKASGTT 2 1 YKFPCNKWIDLDDDEENNSSVTLKCKSADVAASSKAKPV 1 2 ELKPVKYEVTVVTGDEKGAGTDANVSVILYGDNGDTGPRPLKKKFVNLFERNQHDKFTIEALDLGKLTKLHIEHDNKGWGASWLLDRVEVHNVDSNETIIFPCKQWLDKKKGDGQIAKDLLPES* 0 >Monosiga brevicollis (monosiga) Eukaryota|Choanoflagellates XM_001742822 15 PLAT domains MADLPVPTTALQRLQLERSRYSFAAADPPPSFVQRYTSADVLSHRLYAQGLASDVGPGSHSSLGMSRRVELPPYNPLNDPALANYFARKFEWNSTRSVGA GSGSRRSASASLTRTRQPVRGASAHGHTKSRTKSSHRKGHASLEIFTSKRSNPVSKSEKYITVVGTKGRSDAIQLAAPGVHFRAGNKDVFQVNLTGIGKP TKVILENTGTKRTDGWCVSKVVLVKKTDKGTRRYRFSGPVWLSKHHDEMKLKRVLHIDDEDIGSGNGYRIDCYTGDVANAGTDAVATIQLFGSKGQSPMV ELRRSDGQAFQRARVATFTLDDLANLGKLKKLVISHNGHGMASGWFLDKIIVTSLSSNKATVFPCDAWLDRKNGRSKELVARTAAAGAEGMTTFTIRVMT GDRRGAGTDANVQCTLFGEDGESGPHTLNTSRNDFRRGHTDVFAVSSRKIGTLKRLRIWHDNGGAGPAWFLDAVEVVDEASGQTYRFECNRWLAKDEDDG QISRELTCNGDSSWGLKSYKLTIFTGDKRNAGTSANVFCKLVGERGASDNVILENSSKNFQRDRTDIFTVEASDLGSLRHIVLGHDNHGMGAGWYVERFS LEVPSEGKLYNVDVKQWFATDMSDGAIERTFRLDNAETLDIAQRLDWKCTIYTSDVANAGTDANVFMQVYGKKGKTDVVPLKNKSDTFERGQTDELRVQL INVGSLRKLRVWHDNKGMASGWHLDRIVLSRDGEEYIFPCAEWLAVSEGDKEIVRELPATGPNVKKPLQLVEYTVRVATGHARFAGTNADVFVMLTGELG DSGKRALLRSQTNRNKFERGKEDVFTVAAVDLGKLTSVTVGHNNAGTSAGWFLDKIVVLDPRRGEEEEFPCHRWLAVDADDGQIERELVPKMAEHQAAAT TTYIVKIKTGDVRHAGTDANVFVQLFGKTGESTQLKLRNSETYSDAFERNKMDIFKFELLDLGDLSRILVGHDNKGMGAAWFLDYVEVEVPSIRTRWKFP CSRWFSKSQDDGLTEREIYAEKEAGEPMEEDVSAPYLFRFYTSDVAFAGTDANVSVVLYGDEGKTEELVVNNQSDNFERGKADDFKLACKPVGRPSKIRL SAHGGGMSADWHLEKIEVHELGQARIYTFEHNDWLRKGTKAKPFMVELPLRRIETVDDNGREVVEELALDANKRTYRVKVHTGDQKGAGTDANVYVNLHG SLGDSGDRHLKNSLTHTNKFQRKTVDEFDIDAVTLGDINKVKVWHDNAGLGAAWYLEKIEVVDTADDKTYIFPCAQWFAKSMGDGQIARELGVLEEQKPA DFQTKNVGFKYRISVHTSDVKHAGTDANVDIVLYGEKGDTGKIRLAKSETHRDMWERGNCDVFTVSAIELGDLKRVDIMHDGKGVGSGWHLNKVVVDAPQ AGKTWTFMCDAWLDKATDDGTMAKTLYASADAIEEYSAHVPYEIIIKTSDVRNAGTDANVFIDLYGRDQEERDLTAHHEFKDAVKAHFERNLEDRFNVEL PDVGSIYKIRLGHDGKGMSSSWHVASVVVINQRTHERFEFPCDAWLSKDKDDKKLVREFAVGEVKALEGDKVVRRESILSLQEAIYKIHVFTGDIKHAGT DANIYVQIFGDTGDSGEIKLEKSETYRDKFERGHEDIFTHRCLDLGPLRKIKVRSDGKGLMGGDWYLDRVEVHQENDLSEPPVRFVCQDWFKRGKQEGDT LEREITAQVDAAVAMKEATALEDEAARLQRKATTLSRKSTKGSPLRKGTGTTETNAELEEAQRQANIARRKADEAKERAGLAGADDTQLEYKVTVYTGTD TQAGTTANVWLQLFGEKDAPLTPAPPSPSKLSRSGSLFGRRRRASSDAQSVSSSLSAGASGPRETSTGRLQLNNAPADLQSGAKTTFTVTGLDVGELVGL EIGHDDERDKWYLEQVVVEVPKSATHAARRYEFKAGVWLAARADGSTSGSAKGKSKASVRLQPSEIHAGSERILEYTLKVYTASADGAGCTAVPQVQLFG DKHTTEALPLRAGGDILPASVVETQHRVPDLGALLKVRLMVPRGSSWTVEKVEFGRAGQTPITFVGSDGSAVTLGAERLSFDFLPAAAPASSGGKGKRRG STTSSVAVAQQTSYRVYVTTADERGTGTDANVSIILYGAMGDSGEHSLTKSETFDDPFERGNTDVFTLEVPDLGELQRARIWHDGKGMFSSWKLDKIVVV VEATQSRYELPCGQWLSKNKGDKQLTRDLAVASKRVNALTGTYKLEVATDSRAGGGCKGPVRIMLLDAEKNQLPLTLEPPGGEFAPGSVEHLVFDNVMLL GPLTELRIRRKPSGASSRRGAEDDDEDDNEASGASSQSGSAVSPWHLEHIIVKHLQSGQSFVFKGPSKGLSRSRAKLSVHTEATTEEAVAQATKASVAQY EVAVTTGTERGAGTDSNVFVTLFGKNGDSGERALAKSKTFRNMFESGNTDVFDVECQDLGELTKIEVKSDLKGFGAAWQLDKIKVTRTGSQNSWQFKCDQ WFDKKQGAEHTFSVAS*
>LOXHD1_homSap_full dna: each line is an exon ATGATGCCCCAGAAGAAAAGGCGGAGGAAGAAGGACATCGACTTCCTGGCCCTGTACGAAGCGGAGCTGCTGAACTACGCCTCGGAGGACGACGAGGGGGAGCTGGAACACGAGTACTACAAGGCCAGAG TGTATGAAGTGGTCACAGCCACGGGGGATGTTCGCGGTGCAGGGACGGATGCCAATGTCTTCATCACGCTTTTTGGAGAGAATGGGCTCTCTCCCAAGCTCCAGCTCACCAGCAA GAGCAAGTCTGCCTTTGAGAAGGGCAACGTCGATGTGTTCCGGGTGAGAACCAACAATGTGGGCCTCATCTATAAAGTCAG GATTGAGCATGACAACACGGGCTTGAATGCCAGCTGGTACCTGGACCATGTGATTGTGACCGACATGAAGAGGCCTCATCTCCGTTACTACTTCAACTGCAACAACTGGCTGAGCAAGGTGGAAGGTGACCGCCAGTGGTGCCGTGACCTGCTGGCCAGCTTCAACCCCATGGACATGCCCAGAG GTAATAAGTATGAAGTCAAGGTATACACTGGTGATGTAATTGGTGCAGGGACAGATGCTGATGTCTTCATCAATATTTTTGGAGAGTATGGAGACACAG GGGAGCGTAGGCTAGAAAATGAAAAGGACAACTTTGAAAAGGGAGCTGAAGACAGGTTCATCCTGGATGCCCCGGATTTGGGGCAGCTGATGAAGATCAATGTTGGCCACAACAATAAGGGGGGCTCTGCAGGTTGGTTCCTGTCCCAG ATAGTCATTGAAGATATTGGGAACAAAAGAAAATATGACTTCCCCCTTAACCGCTGGCTGGCCTTGGACGAAGACGATGGCAAAATCCAAAGGGATATCTTAGTGGGCGGAGCTGAGACCACAG CTATTACGTATATTGTCACCGTCTTCACTGGGGATGTCCGGGGGGCTGGTACCAAATCCAAAATCTACTTGGTCATGTATGGGGCCAGAGGGAATAAGAACAGTGGGAAAATCTTCCTGGAGGGCGGCGTGTTTGACCGAGGCCGCACGGACATCTTCCACATCGAGCTGGCTGTCCTCCTTAGCCCCCTGAGTCGGGTCTCCGTCGGGCATGGCAATGTGGGTGTCAACAGAGGCTGGTTCTGTGAGAAG GTGGTGATTCTGTGCCCCTTCACTGGTATCCAGCAGACCTTCCCTTGTAGCAACTGGCTGGATGAGAAGAAAGCGGATGGGTTGATTGAGAGGCAGCTCTATGAGATGGTGTCTCTCAGGAAGAAGCGGCTGAAAA AATTCCCTTGGTCCCTGTGGGTCTGGACAACCGACCTAAAGAAAGCTGGTACCAACTCTCCCATCTTCATCCAGATTTATGGGCAGAAGGGGCGGACAGATGAGATTCTCCTGAATCCCAACAACAAGTGGTTCAAACCCGGCATAATCGAGAAGTTTAGG ATTGAGCTCCCGGATCTTGGCAGGTTTTATAAGATTCGAGTATGGCATGATAAAAGGAGTTCTGGTTCTGGATGGCATTTAGAAAGG ATGACCCTGATGAACACTCTGAACAAAGACAAGTACAACTTCAATTGCAACCGCTGGCTGGATGCCAATGAGGATGACAATGAGATAGTGAGGGAAATGACTGCAGAAGGCCCAACAGTGCGCAGGATCATGGGCA TGGCCCGGTACCATGTGACTGTGTGCACAGGTGAACTTGAAGGTGCTGGGACCGATGCCAACGTCTATCTCTGCCTTTTTGGTGATGTGGGGGACACGGGGGAACGGCTGCTCTACAACTGCAGGAATAACACAGACCTGTTTGAAAAGGGCAAT GCTGACGAGTTCACTATCGAGTCTGTCACCATGCGGAATGTGAGGCGGGTGAGGATCAGACACGATGGCAAAGGCTCCGGCAGCGGCTGGTACCTGGACAGAGTGCTGGTGAGAGAGGAGGGGCAGCCTGAGAGCGACAACGTGGAGTTCCCATGTCTCAG GTGGTTGGACAAGGATAAGGATGATGGGCAGCTGGTCCGAGAGTTGCTACCCAGTGACAGCAGCGCGACACTGAAGA ACTTTCGCTATCACATCAGCTTGAAGACTGGGGATGTCTCTGGGGCCAGCACGGATTCTAGAGTCTACATCAAGCTCTATGGGGATAAATCTGACACCATCAAGCAAGTTCTTCTTGTCTCTGACAACAACCTCAAAGACTACTTTGAACGTGGCCGGGTGGATGAGTTCACCCTCGAGACCCTGAACATTGGAAAT ATCAACCGGCTGGTGATTGGGCATGACAGCACTGGCATGCATGCCAGCTGGTTCCTGGGCAGCGTTCAGATCCGTGTGCCCCGTCAAGGCAAGCAGTACACCTTTCCCGCCAACCGCTGGCTGGACAAGAACCAGGCTGACGGGCGCCTGGAGGTGGAGCTGTATCCCAGCGAGGTGGTGGAGATCCAGAAAT TGGTCCACTATGAGGTTGAGATTTGGACAGGAGATGTGGGTGGCGCAGGCACCAGTGCCCGAGTCTACATGCAGATCTATGGAGAGAAAGGCAAGACAGAAGTGCTCTTCCTCTCCAGCCGCTCAAAAGTTTTTGAACGGGCGTCCAAGGACACATTCCAG CTTGAGGCGGCCGACGTGGGCGAGGTCTATAAGCTCCGGCTCGGGCACACGGGCGAGGGCTTTGGGCCCAGCTGGTTCGTGGACACCGTGTGGCTGCGGCACCTGGTGGTGCGGGAGGTGGACCTCACGCCGGAGGAGGAGGCCCGGAAGAAGAAGGAGAAGGACAAGCTGCGGCAGCTGCTCAAGAAGGAGCGGCTGAAGGCCAAGCTGCAGAGGAAGAAGAAGAAGAGGAAGGGCAGCGACGAAGAGGACGAGGGGGAGGAAGAGGAGTCGTCCTCATCAGAGGAGTCCTCGTCAGAGGAGGAGGAGATGGAAGAAGAGGAGGAAGAGGAGGAGTTTGGGCCGGGGATGCAGGAGGTGATTGAGCAGCACAAGTTCGAAGCCCACCGCTGGCTGGCCCGGGGCAAGGAGGACAACGAACTTGTCGTGGAGTTGGTGCCAGCTGGCAAGCCGGGTCCTGAGC GAAACACCTATGAGGTTCAGGTGGTCACGGGGAATGTGCCCAAGGCCGGCACTGATGCTAACGTCTACCTAACCATCTACGGCGAGGAGTATGGAGACACGGGCGAACGACCCCTGAAGAAGTCAGACAAGTCCAACAAATTTGAGCAGGGGCAG ACAGACACCTTCACCATCTATGCCATTGACCTGGGGGCCCTGACCAAGATTCGGATTCGCCACGACAACACAGGCAACAGAGCAGGCTGGTTCCTGGACAGAATAGACATTACTGACATGAACAACGAGATCAC GTACTACTTTCCATGCCAACGTTGGCTGGCAGTGGAGGAAGATGATGGCCAGCTGTCCAGGGAGCTGTTGCCAGTGGATGAGTCCTATGTGCTGCCACAGAGCGAGGAGGGTAGGGGAGGCGGTGACAACAACCCCCTCGACAACCTGGCCCTGGAGCAGAAAG ATAAATCTACCACATTCTCAGTGACCATAAAGACTGGGGTTAAGAAGAATGCGGGCACAGATGCTAATGTCTTCATCACACTCTTTGGCACACAGGATGACACTG GAATGACCCTCCTGAAGTCCTCCAAGACAAACAGCGATAAGTTTGAGAGGGACAGCATTGAAATCTTCACGGTGGAGACGCTGGATCTGGGAGACCTGTGGAAAGTCCGGCTTGGCCATGACAACACAG GCAAGGCCCCAGGCTGGTTTGTAGACTGGGTAGAGGTGGATGCCCCATCTCTTGGGAAGTGCATGACGTTTCCCTGTGGCCGCTGGCTGGCCAAAAACGAAGACGACGGGTCCATCATCAGAGACCTCTTCCATGCAGAGCTTCAGACGAGGCTGTACACACCAT TTGTTCCTTACGAGATCACCCTCTACACCAGTGATGTCTTTGCTGCTGGGACAGATGCCAACATCTTCATCATCATCTATGGCTGCGATGCCGTGTGCACCCAGCAGAAGTATCTGTGTACCAACAAGAGGGAACAGAAGCAGTTCTTTGAGAGGAAGTCTGCCTCCCGCTTCATCGTAGAG TTAGAAGATGTGGGAGAAATCATTGAAAAAATTCGGATTGGCCATAATAACACGGGCATGAATCCTGGGTGGCACTGCTCTCACGTGGACATCCGCAGGCTCCTCCCGGATAAAGAC GGTGCAGAGACCTTGACTTTCCCATGCGATCGGTGGCTTGCCACCTCTGAGGATGACAAAAAGACCATTCGAGAACTGGTTCCATATGACATCTTCACTGAGAAATACATGAAAGATGGGTCCTTACGGCAAGTCTACAAGGAAGTAGAAGAGCCTCTGGACa TTGTGCTGTACTCGGTGCAGATCTTCACAGGGAACATTCCTGGGGCAGGGACGGATGCCAAGGTGTACATCACCATCTATGGAGACCTCGGGGACACTGGGGAGCGATACCTTGGCAAGTCAGAGAACCGGACCAACAAGTTCGAGAGAGGAACG GCTGACACCTTCATCATCGAGGCCGCTGACCTAGGCGTCATCTACAAGATCAAGCTCCGCCATGACAACTCCAAGTGGTGCGCAGACTGGTACGTGGAGAAGGTGGAGATCTGGAATGACACCAACGAGGACGAGTTCCTGTTCCTATGCGGGCGCTGGCTCTCCCTGAAGAAGGAGGATGGGCGACTCGAGAGGCTCTTTTACGAGAAG GAGTACACTGGGGACCGCAGCAGCAACTGCAGCAGCCCTGCTGACTTCTGGGAGATCGCCCTGAGCTCCAAGATGGCCGATGTCGACATCAGCACAGTGACCGGGCCCATGGCTGACTACGTTCAAGAGGGCCCAA TTATTCCCTACTATGTGTCAGTCACCACTGGGAAGCACAAGGACGCGGCCACTGACAGCCGAGCCTTCATCTTTCTCATCGGGGAGGATGATGAACGTAGTAAGCGCATCTGGTTGGACTACCCCCGAGGGAAGAGGGGCTTCAGCCGTGGCTCTGTGGAGGAGTTCTACGTCGCAGGCTTGGATGTGGGCATCATCAAGAAAATAGAG CTGGGCCATGACGGGGCCTCCCCTGAGAGCTGCTGGCTGGTGGAAGAGTTGTGTTTGGCAGTGCCCACCCAGGGCACCAAGTACATGTTGAACTGTAACTGCTGGCTGGCCAAGGACAGAGGCGACGGCATCACCTCCCGTGTCTTCGACCTCTTGGATGCCATGGTGGTGAACATTGGGGTGAAG GTTCTCTATGAAATGACGGTGTGGACAGGGGATGTGGTTGGCGGGGGCACTGACTCCAACATCTTCATGACCCTCTACGGCATCAACGGGAGCACAGAGGAGATGCAGCTGGACAAAAAGAAAGCCAG GTTTGAGCGGGAGCAGAACGACACCTTCATCATGGAGATCCTAGACATTGCTCCATTCACCAAGATGCGGATCCGGATTGATGGCCTGGGCAGTCGGCCGGAGTGGTTCCTGGAGAGG ATCCTACTGAAGAACATGAACACTGGAGACCTGACCATGTTCTACTATGGAGACTGGCTGTCCCAGCGGAAGGGCAAGAAGACCCTGGTGTGTGAAATGTGTGCCGTTATCGATGAGGAAGAAATGATGGAGTGGACCTCCTACACCGTCGCAGTTAAGACCAGCGACATCCTGG GAGCAGGCACTGATGCCAACGTGTTCATCATCATCTTCGGGGAGAACGGGGATAGTGGGACACTGGCCCTGAAGCAGTCGGCAAACTGGAACAAGTTTGAGCGGAACAACACGGACACATTCAACTTCCCTGACATGCTGAGCTTGGGCCACCTCTGCAAGCTGAGGGTCTGGCACGACAACAAAG GGATATTTCCTGGCTGGCATCTGAGCTATGTCGATGTGAAGGACAACTCCCGCGACGAGACCTTCCACTTCCAGTGTGACTGCTGGCTCTCCAAGAGTGAGGGTGACGGGCAGACGGTCCGCGACTTTGCCTGTGCCAACAACAAGATCTGTGATGAGCTGGAAGAGACCA CCTACGAGATCGTCATAGAAACGGGCAACGGAGGCGAAACCAGGGAGAACGTCTGGCTCATCCTGGAGGGCAGGAAGAACCGATCCAAAGAGTTTCTCATGGAAAATTCTTCTAGGCAGCGGGCCTTTAGGAA GGGGACCACAGACACGTTTGAGTTTGACAGCATCTACTTGGGGGACATTGCCTCCCTCTGTGTGGGCCACCTTGCCAGGGAAGACCGGTTTATCCCCAAGAGAGAACTTGCCTGGCATGTCAAGACCATCACCATCACCGAGATGGAGTACGGCAATGT GTACTTCTTTAACTGTGACTGCCTCATCCCCCTCAAGAGGAAGAGGAAGTACTTCAAGGTATTCGAGGTTACCAAGACGACAGAGAGCTTTGCCAGCAAGGTCCAGAGCCTGGTGCCCGTCAAGTACGAAGTCATCGTGACAACAGGCTATGAGCCAGGGGCAGGCACTGATGCCAACGTCTTCGTGACCATCTTTGGGGCCAACGGAGACACAGGCAAGCGGGAGCTGAAGCAGAAAATGCGCAACCTCTTCGAGCGGGGCAGCACAGACCGCTTCTTCCTGGAGACGCTGGAGCTGGGTGAGCTGCGCAAGGTGCGCCTGGAGCACGACAGCAGTGGCTACTGCTCAGGCTGGCTGGTGGAGAAGGTGGAGGTCACCAACACCAGCACCGGCGTGGCCACCATCTTCAACTGTGGCAGGTGGCTGGACAAGAAGCGGGGGGATGGACTCACCTGGAGAGACCTCTTCCCTTCTGTC
Reference PLAT domain sets
>LOXHD1_homSap_LH2:01 chr18: 42343528 Lipoxygenase homology no other domains pdb:3DY5
RVYEVVTATGDVRGAGTDANVFITLFGENGLSPKLQLTSKSKSAFEKGNVDVFRVRTNNVGLIYKVRIEHDNTGLNASWYLDHVIVTDMKRPHLRYYFNCNNWLSKVEGDRQWCRD
>LOXHD1_homSap_LH2:02 chr18: 42343528 Lipoxygenase homology no other domains pdb:3DY5
NKYEVKVYTGDVIGAGTDADVFINIFGEYGDTGERRLENEKDNFEKGAEDRFILDAPDLGQLMKINVGHNNKGGSAGWFLSQIVIEDIGNKRKYDFPLNRWLALDEDDGKIQRDILVGG
>LOXHD1_homSap_LH2:03 chr18: 42343528 Lipoxygenase homology no other domains pdb:3DY5
ITYIVTVFTGDVRGAGTKSKIYLVMYGARGNKNSGKIFLEGGVFDRGRTDIFHIELAVLLSPLSRVSVGHGNVGVNRGWFCEKVVILCPFTGIQQTFPCSNWLDEKKADGLIER
>LOXHD1_homSap_LH2:04 chr18: 42343528 Lipoxygenase homology no other domains pdb:3DY5
FPWSLWVWTTDLKKAGTNSPIFIQIYGQKGRTDEILLNPNNKWFKPGIIEKFRIELPDLGRFYKIRVWHDKRSSGSGWHLERMTLMNTLNKDKYNFNCNRWLDANEDDNEIVREM
>LOXHD1_homSap_LH2:05 chr18: 42343528 Lipoxygenase homology no other domains pdb:3DY5
ARYHVTVCTGELEGAGTDANVYLCLFGDVGDTGERLLYNCRNNTDLFEKGNADEFTIESVTMRNVRRVRIRHDGKGSGSGWYLDRVLVREEGQPESDNVEFPCLRWLDKDKDDGQLVRELLPS
>LOXHD1_homSap_LH2:06 chr18: 42343528 Lipoxygenase homology no other domains pdb:3DY5
FRYHISLKTGDVSGASTDSRVYIKLYGDKSDTIKQVLLVSDNNLKDYFERGRVDEFTLETLNIGNINRLVIGHDSTGMHASWFLGSVQIRVPRQGKQYTFPANRWLDKNQADGRLEV
>LOXHD1_homSap_LH2:07 chr18: 42343528 Lipoxygenase homology no other domains pdb:3DY5
VHYEVEIWTGDVGGAGTSARVYMQIYGEKGKTEVLFLSSRSKVFERASKDTFQLEAADVGEVYKLRLGHTGEGFGPSWFVDTVWLRHLVVREVDLTPEEEARKKKEKDKLR
>LOXHD1_homSap_LH2:08 chr18: 42343528 Lipoxygenase homology no other domains pdb:3DY5
NTYEVQVVTGNVPKAGTDANVYLTIYGEEYGDTGERPLKKSDKSNKFEQGQTDTFTIYAIDLGALTKIRIRHDNTGNRAGWFLDRIDITDMNNEITYYFPCQRWLAVEEDDGQLSRELLPV
>LOXHD1_homSap_LH2:09 chr18: 42343528 Lipoxygenase homology no other domains pdb:3DY5
TTFSVTIKTGVKKNAGTDANVFITLFGTQDDTGMTLLKSSKTNSDKFERDSIEIFTVETLDLGDLWKVRLGHDNTGKAPGWFVDWVEVDAPSLGKCMTFPCGRWLAKNEDDGSIIRDLFHA
>LOXHD1_homSap_LH2:10 chr18: 42343528 Lipoxygenase homology no other domains pdb:3DY5
VPYEITLYTSDVFAAGTDANIFIIIYGCDAVCTQQKYLCTNKREQKQFFERKSASRFIVELEDVGEIIEKIRIGHNNTGMNPGWHCSHVDIRRLLPDKDGAETLTFPCDRWLATSEDDKKTIRELVP
>LOXHD1_homSap_LH2:11 chr18: 42343528 Lipoxygenase homology no other domains pdb:3DY5
VLYSVQIFTGNIPGAGTDAKVYITIYGDLGDTGERYLGKSENRTNKFERGTADTFIIEAADLGVIYKIKLRHDNSKWCADWYVEKVEIWNDTNEDEFLFLCGRWLSLKKEDGRLERLFYEK
>LOXHD1_homSap_LH2:12 chr18: 42343528 Lipoxygenase homology no other domains pdb:3DY5
IPYYVSVTTGKHKDAATDSRAFIFLIGEDDERSKRIWLDYPRGKRGFSRGSVEEFYVAGLDVGIIKKIELGHDGASPESCWLVEELCLAVPTQGTKYMLNCNCWLAKDRGDGITSRVFD
>LOXHD1_homSap_LH2:13 chr18: 42343528 Lipoxygenase homology no other domains pdb:3DY5
VLYEMTVWTGDVVGGGTDSNIFMTLYGINGSTEEMQLDKKKARFEREQNDTFIMEILDIAPFTKMRIRIDGLGSRPEWFLERILLKNMNTGDLTMFYYGDWLSQRKGKKTLVC
>LOXHD1_homSap_LH2:14 chr18: 42343528 Lipoxygenase homology no other domains pdb:3DY5
TSYTVAVKTSDILGAGTDANVFIIIFGENGDSGTLALKQSANWNKFERNNTDTFNFPDMLSLGHLCKLRVWHDNKGIFPGWHLSYVDVKDNSRDETFHFQCDCWLSKSEGDGQTVRDFAC
>LOXHD1_homSap_LH2:15 chr18: 42343528 Lipoxygenase homology no other domains pdb:3DY5
TTYEIVIETGNGGETRENVWLILEGRKNRSKEFLMENSSRQRAFRKGTTDTFEFDSIYLGDIASLCVGHLAREDRFIPKRELAWHVKTITITEMEYGNVYFFNCDCLIPLKRKRKYFKVF
>LOXHD1_homSap_LH2:16 chr18: 42343528 Lipoxygenase homology no other domains pdb:3DY5
VKYEVIVTTGYEPGAGTDANVFVTIFGANGDTGKRELKQKMRNLFERGSTDRFFLETLELGELRKVRLEHDSSGYCSGWLVEKVEVTNTSTGVATIFNCGRWLDKKRGDGLTWRDLFPS
>PKD1L2_homSap_LH2:01 chr16: 79691985 polycystin 1-like 2 signal...GPS-TM-LH2-TM pdb:3CWZ
YHYLVTVYTGHRRGAATSSKVTVTLYGLDGEREPHHLADPDTPVFERGAVDAFLLSTLFPLGELRSLRLWHDNSGDRPSWYVSRVLVYDLVMDRKWYFLCNSWLSINVGDCVLDKVFPVA
>PKDREJ_homSap_LH2:01 chr22: 45030224 receptor for egg jelly-like signal...GPS-TM-LH2-TM pdb:3CWZ
CYLVTIFTGSRWGSGTRANVFVQLRGTVSTSDVHCLSHPHFTTLYRGSINTFLLTTKSDLGDIHSIRVWHNNEGRSPSWYLSRIKVENLFSRHIWLFICQKWLSVDTTLDRTFHVT
>PKD1L1_homSap_LH2:01 chr7: 47780815 polycystin-1L1 GPS-TM-LH2-TM pdb:3CWZ
QLYAVVIDTGFRAPARLTSKVYIVLCGDNGLSETKELSCPEKPLFERNSRHTFILSAPAQLGLLRKIRLWHDSRGPSPGWFISHVMVKELHTGQGWFFPAQCWLSAGRHDGRVERELTC
>PKD1_homSap_LH2:01 chr16: 2078712 polycystin 1 GPS-TM-LH2-TM pdb:3CWZ
FKYEILVKTGWGRGSGTTAHVGIMLYGVDSRSGHRHLDGDRAFHRNSLDIFRIATPHSLGSVWKIRVWHDNKGLSPAWFLQHVIVRDLQTARSAFFLVNDWLSVETEANGGLVEK
>DENND5B_homSap_LH2:01 chr12: 31426424 DENN/MADD domain 5B RUN-LH2-RUN pdb:3CWZ
RSVIIPIKKLSNAIITSNPWICVSGELGDTGVMQIPKNLLEMTFECQNLGKLTTVQIGHDNSGLLAKWLVDCVMVRNEITGHTYRFPCGRWLGKGIDDGSLER
>DENND5A_homSap_LH2:01 chr11: 9116949 RAB6 interacting protein 1 RUN-LH2-RUN pdb:3CWZ
HILIVPSKKLGGSMFTANPWICISGELGETQIMQIPRNVLEMTFECQNLGKLTTVQIGHDNSGLYAKWLVEYVMVRNEITGHTYKFPCGRWLGKGMDDGSLER
>ALOX5_homSap_LH2:01 chr10: 45189635 arachidonate 5-lipoxygenase NH2-LH2-lipox pdb:2P0M
PSYTVTVATGSQWFAGTDDYIYLSLVGSAGCSEKHLLDKPFYNDFERGAVDSYDVTVDEELGEIQLVRIEKRKYWLNDDWYLKYITLKTPHGDYIEFPCYRWITGDVEVVLRDG
>ALOXE3_homSap_LH2:01 chr17: 7939943 arachidonate lipoxygenase 3 NH2-LH2-lipox pdb:2P0M
AVYRLCVTTGPYLRAGTLDNISVTLVGTCGESPKQRLDRMGRDFAPGSVQKYKVRCTAELGELLLLRVHKERYAFFRKDSWYCSRICVTEPDGSVSHFPCYQWIEGYCTVELRPG
>ALOX15_homSap_LH2:01 chr17: 4480963 arachidonate 15-lipoxygenase NH2-LH2-lipox pdb:2P0M
GLYRIRVSTGASLYAGSNNQVQLWLVGQHGEAALGKRLWPARGKETELKVEVPEYLGPLLFVKLRKRHLLKDDAWFCNWISVQGPGAGDEVRFPCYRWVEGNGVLSLPEG
>ALOX12B_homSap_LH2:01 chr17: 7916679 arachidonate 12R-lipoxygenase NH2-LH2-lipox pdb:2P0M
ATYKVRVATGTDLLSGTRDSISLTIVGTQGESHKQLLNHFGRDFATGAVGQYTVQCPQDLGELIIIRLHKERYAFFPKDPWYCNYVQICAPNGRIYHFPAYQWMDGYETLALREA
>ALOX12_homSap_LH2:01 chr17: 6840108 arachidonate 12-lipoxygenase NH2-LH2-lipox pdb:2P0M
GRYRIRVATGAWLFSGSYNRVQLWLVGTRGEAELELQLRPARGEEEEFDHDVAEDLGLLQFVRLRKHHWLVDDAWFCDRITVQGPGACAEVAFPCYRWVQGEDILSLPEG
>LOXHD1_schMan_PLAT:01 Schistosoma mansoni (roundworm) Bilatera; Platyhelminthes; Trematoda XP_002576380 from contig NS_000200
TLYKVVLTTADVPGAGTSAQIYITLKGEWGSSTRQKLRKEKVPTRNLRFYFYPGSTNTFSVVSPDLGGLHSVFIEHDSLRKSDSWLLESVQVFHPLTKKRYMFMCNHWFSLYKEDGLIARELF
>LOXHD1_schMan_PLAT:02
TKYSIVTVTGDQEGSGTNSKVYITIYGRTGITPRIELSQENKSTGKDILCAPFGRGTSTKFIVKAPNVGAITNIRIKQDESGNEPHWFLERVVVTDMSYPQWTYYFHLSCWLSSKYGDGKSCR
>LOXHD1_schMan_PLAT:03
TEYKLTFYTSDQKDAGTTGEVYVKLYGEKDSSREIWVNSINQKNRRQPTSYHFTRGSTVEVYLPPCPQLGEITKLKVGHNRAGSSPSWFLNKVIVDDLRMNRVFEFPCYAWITK
>LOXHD1_schMan_PLAT:04
IPLEIRLYTGDVPNAGTTAKVYLHQKTTSENKEDVYTTPYIWLEDGNYDRNGVTLFSIDLPVPKFISPLSQLVIGHDNTGHSPSWFFDKVQIYCPLNGVEQTFLYRKWLTSSKPGMKVEQILCEE
>LOXHD1_schMan_PLAT:05
IPWEISVKTSPIANSYVTAAVSIIIFGSKDKTMKIQLDRSNLEVSSEWTKVSGNEVVEMFRPNVESKFRIYIKDIGIPCKLRIQHDNKGSNPNWHLQEIVLTNLRTHEQYEFYCNRWLSTREDDGSILREIPAK
>LOXHD1_schMan_PLAT:06
YIIQIFTGNKPNSGTNANIFINIFGEKGDCGERWLGRSVNRNSELFQQNQMDEFVIEAVQLGSINKICIGHEERSPGYGWYLAKIVLTIKENPKYKLTFECYRWFDVGEDDGQIVRELFA
>LOXHD1_schMan_PLAT:07
IAYNVTVLTGTCRNAGTVANVFVHLYGLQGESKDMQLKHKETEITKFEAGKSEEFILACGKLGEIKKIKIWHDSIAPHSGWFLDEVVVIDYFLGRRYVFHVNRWLAKNEADGLISLDLQPT
>LOXHD1_schMan_PLAT:08
SYEVIVKTGGRKYAGTDSHVYITMFGINSESKEYHLANSKTHNNKFEQNHEDLFNLNAVGLGSLKKIRVRHTNTGIAPGWFLDYILIRESIEQKKMEYFFPCYQWLSATRLDGLVIREL
>LOXHD1_schMan_PLAT:09
TIYHVRIHTGSQSNISSDANVQIQLYGDKDFTGNIRLYKAYYGKEDILVNKFQAGQIANFIVKAINIGNIKKIRIEHDTAVGSARWFVERVEVEARKLGLLWKFECNR
>LOXHD1_schMan_PLAT:10
TNYQIKTFTSDISGAETTSSVYIQLYGNDSLPSSIRRLHQNNDSEQRFQRNKIDTFYVELEELQEPFSKLRIWHNDKGSSTDWHLNKVEIRKIKTNQYVFVTYIFPCNKWLSRNMDQAALERELIPS
>LOXHD1_schMan_PLAT:11
NTYEVRITTGDKAYAGTDASVYITLFGENGDSGERKLTKSLTHRNKFERGQTDVFQLEIVDLGKINKVRIRHDNSGVNPSWYLSTIEVFNISKSQSVHGLKDQPNETHHLVKYIFNCEAWLSTEHDEKVLDR
>LOXHD1_schMan_PLAT:12
IPYLITVITGSDRKAGTPGPVWISCVDKDKMNSEKFILCDCYNRTMLKRGTTRYFRFAGVKLNDLTEIQVGNDAPESPSMGWYIQSLYLSFPTIGKMYLFDCKEWLSTNRGNKK
>LOXHD1_schMan_PLAT:13
ITYHLKITTANVKRAGTDCSINLQIFGTNGVTNCYILEKTSNRFAQGITDNISLEMEDVGKLLKMRIGHDNQGKNKHWNLSCVEVTVANTNQLYRFVYDDWLSLTYGKRKSLWADLPAM
>LOXHD1_schMan_PLAT:14
IFVKTGNMPASSTDANVYVQLFGEYGDSGEILLKQTVSNQKPFQNNSIDHFKIPSILKLGNLARCRIWHDNKGSSPNWYCEWLEVKEVLIPGEKNLACNWKFAFNKWLSVSDDNKQLLRDAPCS
>LOXHD1_schMan_PLAT:15
VYEVVIETGNLKDSGTTCDAWIILEGKHGRSPKLELVNQVGNPILQINQMNTFQLPSFPLGDLETIRLGIQERNINKQTNPNDIQSQKWFCEKVSIKDPVSKRTYIFTIHQWLS
>LOXHD1_schMan_PLAT:16
VMYKVSIYTGTKGCANTDANIFITMFSTTPGLNSGRIALKRENNNLFDRKQLDEFYVESIDLESIQRIIIEHDNTGVSPDWYLDKVLITNQTNNQIHLFQCYQWISKKKGDCRLWKELLVS
Reference PLAT:07 domains
The seventh PLAT domain (human numbering) is unique among PLAT domains in having a large expansion in the middle of the domain. It is a unique signature of the LOXHD1 orthology class that would distinguish it from other poly-PLAT domain concatenates of independent origin. That issue is completely hypothetical because no other protein in any vertebrate genome contains more than a single PLAT domain. Indeed, the LOXHD1 gene is single-copy in all species that retain it (including teleost fish). Thus even with very ancient eukaryotic divergencs, the corresponding genes can be taken as straightforward human orthologs.
These other proteins consist of 10 genes, namely four polycystins (PKD1L2, PKDREJ, PKD1L1, PKD1), two RAB6 (DENND5B, DENND5A), and five arachidonate 5-lipoxygenases (ALOX5, ALOXE3, ALOX15, ALOX12B, ALOX12) plus three bogus misannotations (AK310228, FLJ00285 and NPIP) found in many database compilations. In these other genes the PLAT domain occurs in the context of GPS-TM-PLAT-TM (polycystins), RUN-LH2-RUN (RAB6), and N-terminally (lipoxygenases).
Presumably a loop out of the beta barrel, it consists of an extremely high AG content at the dna level and encodes charged residues, primarily lysine K and glutamate E residues. These are asymmetrically distributed with the lysines N-terminal and the glutamates distal. The region is called the KE loop of PLAT:07 in this article.
Despite minimal net charge, this region may not be able to interact with (hydrophobic) membranes. Yet the split PLAT domain retains the potential to form a standard beta barrel. The region is quite variable even within mammals and must be hand-annotated to ensure accuracy (because its quasi-repetitive nature wreaks havoc with alignment). This section uses comparative genomics within eukaryotes to answer the following questions:
- when did the high GA section arise in phylogenetic time? [early eukaryotes]
- what is its source? [internal expansion, replication slippage]
- did it arise from intron or retroposon retention? [species lacking the repeat also have a single exon]
- can the boundaries of 7th PLAT domain be better located? [conservation on both sides of the KE region allow definition of the PLAT domain segments]
- is the 44-species alignment accurate in this region? [corrections are needed when longer than human, eg platypus]
- does the KE domain disrupts PLAT function? [evidently not: the split domain is highly conserved and can form a normal beta barrel]
- is it a mammalian innovation or an unwanted intrusion? [this ancient expansion has variable expansion and contraction in different clades]
homSap LEAADVGEVYKLRLGHTGEGFGPSWFVDTVWLRHLVVREVDLTPEEEARKKKEKDKLRQLLKKERLKAKLQRKKKKRKGSDEEDEGEEEESSSSEESSSEEEEMEEEEEEEE FGPGMQEVIEQHKFEAHRWLARGKEDNELVVELVPAGKPGPE 1 ornAna LEATDVGEIYKVRLGHSGEGFGSGWFIESLVLKRLVLKEVEPNPEEEKRKAKERERAREQRRKERLKAKQQRKKKKKMKKSSDDEDSEAEDSEEEEGSSEEESSSSSSEEEVEEEFGPGIKEVIDVYKFEAHRWLARDEDDKELIVELEPANRPGPE 1 galGal VEAADVGKIYKIRIGHDGKGIGDGWFLESVTLKRLATKMDETD KKKKKKKKKSEEEEEEEE TKVEEVMDVYTFVAHRWLAKDEGDKELVVELVPDGESELE 1 taeGut REAADVGKIYKIRIGHDGTGIGDGWFLESVTLKRLATKTEGSD KKKKKKKKSEEEETKE EEGMDVYTFVAHRWLAKDEGDKELVVELVPDGESDLE 1 anoCar VEAVDVGKVYKIRIGHDGKGFGDGWFLDSVVVKKLPTKVP KKKKKKKKKKTPEEEEAEE GPGIMEVYNFTPCRWLASDEEDKELVVELVPDEGSELE 1 oryLat IEALDVGKIYKIRIYHDGSGIGDGWFLETVDIKRLTMALVQVEV KKEEAPKKDKKKDKKKKKKEEEEVEIIEE MQEVVETFTFTCNRWLARDEEDGEIVVELLTEENEDLE 1 gasAcu IEAKDVGKIFKIRIGHDGSGIGSGWFLETVDVKRLILALVPKEK KKEDKKKKKKKKEDVDEEGGEE MQEVVLTYSFPCSRWLAGGEEDGELVVELLPDDAKELE 1 takRub IEAKDVGKIFKIRIGHDGLGIGSGWFLEKVYVKHLIMALVPREN KKDDKKKKKKKKKDKEDEEEVGGEE MQEVVVTYHFPCSRWLASGEDDDDLVVELLPEDAEELE 1 petMar VEAMDVGKVVKLRVGHDNSGMGSGWFLDSIVIRRLRQSSPHRPQPVDA EEDEDEEDDEEAEDED VQTYTFPCKRWLARDEDDGEIVRELLPQDCAEME 1 sacKow IEAADVAMLTKIRIGHDNSGRSAGWYLERVIIERFPPKRKM KRKRSGTPRRRGEYDEEDYDD IPETNVVNFVCNRWFAKDEEDHQIVRELLPTDEEALKGH 1 triAdh VECEDVGKLRKLRIGHDSAGMGSAWFLDKVYVRRLPPKSGKKSKET DEREEETAKKDADEPEKLDE NNYLFVANRWLSKEEGDRQTVIEISPVGVDGALA 1 monBre LACKPVGRPSKIRLSAHGGGMSADWHLEKIEVHELGQAR IYTFEHNDWLRKGTKAKPFMVELPLRRIETVDDN 1 >PLAT:07_homSap Homo sapiens (human) 2 LVHYEVEIWTGDVGGAGTSARVYMQIYGEKGKTEVLFLSSRSKVFERASKDTFQ 0 0 LEAADVGEVYKLRLGHTGEGFGPSWFVDTVWLRHLVVREVDLTPEEEARKKKEKDKLRQLLKKERLKAKLQRKKKKRKGSDEEDEGEEEESSSSEESSSEEEEMEEEEEEEE FGPGMQEVIEQHKFEAHRWLARGKEDNELVVELVPAGKPGPE 1 >PLAT:07_monDom Monodelphis domestica (opossum) 2 LIHYEVEIVTGDMGNASTSARVYMQIYGEEGKTEVLFLKSRSKVFQRGNTDTFQ 0 0 LQEAEVTPEEEARKKKEMDKLRQLMRKERLKAKEEQKRKRKLAKGSDVEDEEESSSEESSEESEEETEIEEEDEE FQEVVDVYRFHAGRWLATDEEDKDLDMELAPSGRSGPQR 1 >PLAT:07_ornAna Ornithorhynchus anatinus (platypus) 2 LIHYEVEIVTGDMGYAGTNARVYMQIYGELGKTEVLHLTSRTNVFEQGATDTFQ 0 0 LEATDVGEIYKVRLGHSGEGFGSGWFIESLVLKRLVLKEVEPNPEEEKRKAKERERAREQRRKERLKAKQQRKKKKKMKKSSDDEDSEAEDSEEEEGSSEEESSSSSSEEEVEEEFGPGIKEVIDVYKFEAHRWLARDEDDKELIVELEPANRPGPE 1 >PLAT:07_galGal Gallus gallus (chicken) 2 LINYEVSVVTGDVRAAGTNAKVFMQIYGETGKTELIILENRSNNFERGATDIFE 0 0 VEAADVGKIYKIRIGHDGKGIGDGWFLESVTLKRLATKMDETDKKKKKKKKKSEEEEEEEETKVEEVMDVYTFVAHRWLAKDEGDKELVVELVPDGESELE 1 >PLAT:07_taeGut Taeniopygia guttata (finch) 2 LINYEVSVVTGDVRAAGTNAKVFMQIYGETGKTELIILENRSNNFERGATDIFK 0 0 REAADVGKIYKIRIGHDGTGIGDGWFLESVTLKRLATKTEGSDKKKKKKKKSEEEETKEEEGMDVYTFVAHRWLAKDEGDKELVVELVPDGESDLE 1 >PLAT:07_anoCar Anolis carolinensis (lizard) 2 LINYEVCVATGDVRNAGTNANVFMQIYGELGKTELLILKNRSNNYERGASETFR 0 0 VEAVDVGKVYKIRIGHDGKGFGDGWFLDSVVVKKLPTKVPKKKKKKKKKKTPEEEEAEEGPGIMEVYNFTPCRWLASDEEDKELVVELVPDEGSELE 1 >PLAT:07_oryLat Oryzias latipes (medaka) 2 VINYEIAVVTGDVRAGGTNASVFCQIYGEEGKTEVLNLKSRSNNFERGTTEIFK 0 0 IEALDVGKIYKIRIYHDGSGIGDGWFLETVDIKRLTMALVQVEVKKEEAPKKDKKKDKKKKKKEEEEVEIIEEMQEVVETFTFTCNRWLARDEEDGEIVVELLTEENEDLE 1 >PLAT:07_gasAcu Gasterosteus aculeatus (stickleback) 2 VINYEVTVVTGDVTFAGTNARVIIQIYGDKGKTEVIILESRSNNYERNTTEIFK 0 0 IEAKDVGKIFKIRIGHDGSGIGSGWFLETVDVKRLILALVPKEKKKEDKKKKKKKKEDVDEEGGEEMQEVVLTYSFPCSRWLAGGEEDGELVVELLPDDAKELE 1 >PLAT:07_takRub Takifugu rubripes (fugu) 2 VINYEVTVVTGDVTFAGTNAKVFVQIYGDKGKTEVIMLESRSNNYERNAMEIFK 0 0 IEAKDVGKIFKIRIGHDGLGIGSGWFLEKVYVKHLIMALVPRENKKDDKKKKKKKKKDKEDEEEVGGEEMQEVVVTYHFPCSRWLASGEDDDDLVVELLPEDAEELE 1 >PLAT:07_petMar Petromyzon marinus (lamprey) 2 GINYQVSVHTGNVRSAGTNANVFIQLYGDMGKTEVHNLRNRSDNFERASNDIFK 0 0 VEAMDVGKVVKLRVGHDNSGMGSGWFLDSIVIRRLRQSSPHRPQPVDAEEDEDEEDDEEAEDEDSEEVQTYTFPCKRWLARDEDDGEIVRELLPQDCAEME 1 >PLAT:07_sacKow Saccoglossus kowalevskii (acornworm) Bilateria|Deuterostomia|Hemichordata 2 GIPYEITVITGDKSGAGTDAQVFIRMYGLKGKTDEFILNNRTDNFERGMTDKFK 0 0 IEAADVAMLTKIRIGHDNSGRSAGWYLERVIIERFPPKRKMKRKRSGTPRRRGEYDEEDYDDIPETNVVNFVCNRWFAKDEEDHQIVRELLPTDEEALKGH 1 >PLAT:07_strPur Strongylocentrotus purpuratus (sea_urchin) 2 GIPYEITVITGDKSGAGTDANVFLTMYGDDGAKTEEFSLRNRTDNFEKNMTDKFK 0 >PLAT:07_triAdh Trichoplax adhaerens (placozoan) Eukaryota|Metazoa|Placozoa 2 KIPYQISVHTGDIRHAGTDSNVFAVIYGENGKTEELKLRNKSDNFERGQVDVFK 0 0 VECEDVGKLRKLRIGHDSAGMGSAWFLDK 00 VYVRRLPPKSGKKSKETDEREEETAKKDADEPEKLDENNYLFVANRWLSKEEGDRQTVIEISPVGVDGALA 1 >PLAT:07_monBre Monosiga brevicollis (choanoflagellate) Eukaryota|Choanoflagellida 2 PMEEDVSAPYLFRFYTSDVAFAGTDANVSVVLYGDEGKTEELVVNNQSDNFERGKADDFK 0 0 LACKPVGRPSKIRLSAHGGGMSADWHLEKIEVHELGQARIYTFEHNDWLRKGTKAKPFMVELPLRRIETVDDNGREVVEELALDANKRTYR 1