Opsin evolution: RPE65
Introduction to the RPE65 gene family
This gene family is critical to the metabolism of provitamin A dietary carotenoids and thus to retinoic acid signaling and the recycling of all-trans retinal needed to replenish vertebrate ciliary opsins after exposure to light. RPE65, as the illegimate gene name suggests, is a protein of 65 kdaltons found in the retinal pigment epithelium adjacent to photoreceptor layers.
In vertebrates including human, RPE65 has two full-length paralogs on different chromosomes, BCMO1 (ß-carotene-15,15'-oxygenase cleaving symmetricaly to yield two retinals) and BCO2 (ß-carotene-9',10'-oxygenase cleaving asymmetricaly to yield a retinoic acid precursor and also cleaving lycopene).
The nomenclature of these three genes violates the spirit and letter of international agreement for naming human gene families, yet the responsible committee has approved the names! While an improvement over monstrosities such as B-DIOX-II and BCDO2 beta-carotene dioxygenase (for a mono-oxygenase!), it should be noted that RPE65 is 61 kd and expressed in a great many other tissues (such as skin) beyond retinal pigment epithelium (where BCMO1 also occurs). A more appropriate nomenclature would be BCO1S, BCO2A, and BCO3R, where the letters denote symmetric, asymmetric, and retinal.
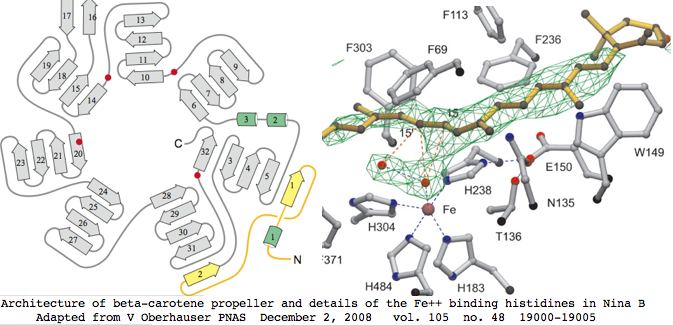
This gene family is readily tracked back to bacteria where a 3D structure has been determined that suffices to model the entire family in all species. Four invariant histidines that hold the catalytic ferrous iron lie on the axis of a seven-bladed beta-propeller fold. The Fe2+ is accessible to carotenoids via a long nonpolar tunnel capable of promoting cis-trans double bond conversions. RPE65 shares all these features even though it is merely an isomerase.
Since this complex structure consists of a single coherent domain fold, alternative coding splices can be dismissed out of hand as transcriptional noise not leading to functional enzyme. The domain structure implies all homologs from all species must be full length or around 550 amino acids, which is validated below over tens of billions of years of evolutionary branch length. That conclusion must be qualified to the extent that N- and C-terminal extensions could be trimmed post-translationally and small indels not affecting the propellers might be tolerated.
RPE65 is a well-known disease gene for retinitis pigmentosa and type II Leber congenital amaurosis. The other two are not currently associated with human disease but have various consequences in knockout mice consistent with enzymatic expectations.
The confused history of RPE65
RPE65 was the subject of recent studies with serious experimental and interpretive error, such as attributing its enzymatic activity to LRAT (ecithin:retinol acyl transferase), assigning palmitolyation to various cysteine residues that did not occur, and attributing its (non-passaging) membrane association to these lipids when in fact that is intrinsic, and misunderstanding enzyme mechanism.
While this never made sense in view of full-length alignment of RPE65 with its two well-studied paralogous carotenoid oxygenases, the most interesting aspect of the debacle is the light it sheds on the multi-year delay cycle in updates at primary hub databases such as RefSeq, SwissProt, and OMIM. None of these have been corrected 18 months later. It's not clear when they will ever be revisited. Consequently hundreds of derived databases such as UCSC perpetuate serious misunderstandings about this important but straighforward disease gene and the visual pigment cycle in vertebrates.
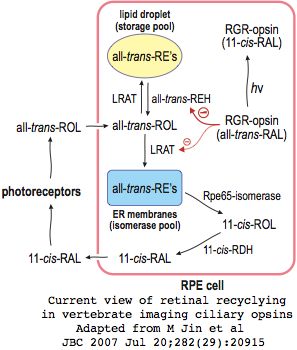
The current view of retinal cycling in vertebrate eyes envisions the primary photosensitive event as isomerization of opsin-bound 11-cis retinal to all-trans retinol. After importation to the adjacent retinal pigment epithelia, that is esterified by LRAT palmitolyation and subsequently isomerized by RPE65 to 11-cis retinol, which must then be oxidized to 11-cis retinal by RDH5 before it can be re-exported back to the retina and re-form its Schiff base in a newly recharged opsin molecule. This system seems quite bizarre in comparison to self-recharging insect melanopsins.
Reference gene collection: non-teleosts
Here the primary focus is obtaining reliable full length genes for the three members of this gene family from early diverging species (rather than standard teleost sequences more closely related to human), the idea being to possibly correlate gene expansions of the RPE65 family with the origin of imaging vision in deuterostomes. However partial sequences are also of valuable for purposes of establishing paralog numbers and presence.
Here it must be immediately noted that many Gnomon predicted sequences at GenBank are so deeply flawed that their uncritical use would hopelessly taint any comparative genomics effort. Without transcripts and given the rates of divergence, it is quite difficult to recover full length genes by blast into incomplete and sometimes garbled assemblies. Of the early deuterostome invertebrates, only Ciona has an adequate (tileable) set of transcripts vis-a-vis this gene family. Yet tunicates have lost one family member altogether, BCO2.
This gene family is quite unusual in that new members are quite difficult to classify by blast clustering relative to a reference collection. The match qualities tend to be quite similar, possibly attributable to gap issues and regions of uncertain and unpersuasive alignment. Here it is best to classify by individual exons because each paralog is intronated distinctively.
Because intronation patterns are strongly conserved from human back to pre-bilaterans (eg cnidarian and placamorpha), the fact that the three paralogs are intronated quite differently implies gene duplication and divergence occured very early, prior to the main era of intron establishment in early pre-metazoan eukaryotes. This is consistent with full length counterparts with conserved 3D structure already existing in bacteria.
This in turn implies that loss of one or more family members has occured in many lineages. For example, arthropods (notably many insect species) contain but a single gene copy (denoted NinaB), lophotrochozoan genomes have none (despite ciliary opsins), and tunicates have two. Gene expansions have also occured in species such as Nematostella yet no expansion occured in deuterostomes despite supposed 1R and 2R whole genome expansions.
>RPE65_homSap length=547 14 exons 0 MSIQ 21 VEHPAGGYKKLFETVEELSSPLTAHVT 1 2 GRIPLWLTGSLLRCGPGLFEVGSEPFYHLFDGQALLHKFDFKEGHVTYHRR 2 1 FIRTDAYVRAMTEKRIVITEFGTCAFPDPCKNIFSR 2 1 FFSYFRGVEVTDNALVNVYPVGEDYYACTETNFITKINPETLETIKQ 0 0 VDLCNYVSVNGATAHPHIENDGTVYNIGNCFGKNFSIAYNIVKIPPLQA 1 2 DKEDPISKSEIVVQFPCSDRFKPSYVHS 21 FGLTPNYIVFVETPVKINLFKFLSSWSLWGANYMDCFESNETMG 0 0 VWLHIADKKRKKYLNNKYRTSPFNLFHHINTYEDNGFLIVDLCCWKG 21 FEFVYNYLYLANLRENWEEVKKNARKAPQPEVRRYVLPLNIDK 0 0 ADTGKNLVTLPNTTATAILCSDETIWLEPEVLFSGPRQ 1 2 AFEFPQINYQKYCGKPYTYAYGLGLNHFVPDR 0 0 LCKLNVKTKETWVWQEPDSYPSEPIFVSHPDALEEDD 1 2 GVVLSVVVSPGAGQKPAYLLILNAKDLSEVARAEVEINIPVTFHGLFKKS* 0 >BCMO1_homSap length=547 11 exons 51,666 bp 0 MDIIFGRNRKEQLEPVRAKVT 1 2 GKIPAWLQGTLLRNGPGMHTVGESRYNHWFDGLALLHSFTIRD 1 2 GEVYYRSKYLRSDTYNTNIEANRIVVSEFGTMAYPDPCKNIFSK 2 1 AFSYLSHTIPDFTDNCLINIMKCGEDFYATSETNYIRKINPQTLETLEK 0 0 VDYRKYVAVNLATSHPHYDEAGNVLNMGTSIVEKGKTKYVIFKIPATVP 1 2 EGKKQGKSPWKHTEVFCSIPSRSLLSPSYYHSFGVTENYVIFLEQPFRLDILKMATAYIRRMSWASCLAFHREEK 0 0 TYIHIIDQRTRQPVQTKFYTDAMVVFHHVNAYEEDGCIVFDVIAYEDNSLYQLFYLANLNQDFKENSRLTSVPTLRRFAVPLHVDK 0 0 NAEVGTNLIKVASTTATALKEEDGQVYCQPEFLYE 1 2 GLELPRVNYAHNGKQYRYVFATGVQWSPIPTK 0 0 IIKYDILTKSSLKWREDDCWPAEPLFVPAPGAKDEDD 1 2 GVILSAIVSTDPQKLPFLLILDAKSFTELARASVDVDMHMDLHGLFITDMDWDTKKQAASEEQRDRASDCHGAPLT* 0 >BCO2_homSap length=579 12 exons alt leader peptide not shown 0 MGNTPQKKAVFGQCRGLPCVAPLLTTVEEAPRGISARVWGHFPKWLNGSLLRIGPGKFEFGKDK 2 1 YNHWFDGMALLHQFRMAKGTVTYRSKFLQSDTYKANSAKNRIVISEFGTLALPDPCKNVFERFMSRFELPGKAA 1 2 AMTDNTNVNYVRYKGDYYLCTETNFMNKVDIETLEKTEK 0 0 VDWSKFIAVNGATAHPHYDLDGTAYNMGNSFGPY 1 2 GFSYKVIRVPPEKVDLGETIHGVQVICSIASTEKGKPSYYHSF 1 2 GMTRNYIIFIEQPLKMNLWKIATSKIRGKAFSDGISWEPQCNTRFHVVEKRTGQ 0 0 LLPGRYYSKPFVTFHQINAFEDQGCVIIDLCCQDNGRTLEVYQLQNLRKAGEGLDQ 0 0 VHNSAAKSFPRRFVLPLNVSLNAPEGDNLSPLSYTSASAVKQADGT 0 0 IWCSHENLHQEDLEKEGGIEFPQIYYDRFSGKKYHFFYGCGFRHLVGDSLIKVDVVNKTLK 0 0 VWREDGFYPSEPVFVPAPGTNEEDGGVILSVVITPNQ 0 0 NESNFILVLDAKNFEELGRAEVPVQMPYGFHGTFIPI* 0 >RPE65_cioInt 40% but about equally similar, slightly different intronation 0 MFAIQRRPFFAVFRNFNKMSAPSKTKSYVKLLQKAEERANAECVVT 1 2 GCIPEWLNGDVLRNGPAEFDIGPDTFKHWFDGHALLHK 2 1 FSMFEGKVTYSSKFLRSGTYKTNHENSRIIIGEFGTASRPDPCKNMFSR 2 1 FFTNFVEIAPRSDNANVSVAQLGEAYYAITDGPTAYGFDPETLETKNLITDCGPANMTVTAAHPHY 1 2 DRNGDYLNLGTTFGRTPHYHVIKVPAAKMTSPDPMNELEVFMKFPSTTSNASYHHS 2 1 FGLSENWIIFHEQPFSFSTPKLLIGLKLWNPILSSFYEDKQTIS 0 0 FHIINKTTGEKIATKYEARGMFCFHHINAYETKENDGKRFIVVDMCGSDRSLVWLL 2 1 GLDTLLDEEAHDKVVSNLDEKYLTRPRRIVIPLDISSDTPN 1 2 DTNLVTIPGCKATAMLNKSGVVSLTYELLVPDDFPNTELGIELPRINYDGYNGREYK 2 1 FIYAISSEYILPSHLVKINVETKEIKYWKEK 0 0 DKYTSEPIFVPRPGSQDEDDGVVLSTVISPTDDKTFLLILDGQSFKEIARAE 0 0 IETKMSYPLHGLFSK* 0 >BCMO1_cioInt 525 aa tiled transcripts agree with genomic 0 MDFPVSAFPHLTALATTKNIEYAEAVQGKVQ 1 2 GEVPSWLNGSWYRNGPGVVHFREESVKHWFDGMALARK 2 1 FCIEDGKVSYMSRLVDGESLQKNTAAGRVVVAEFGTTTHSEGFLGR 2 1 VKSALTMPEFTDNCLINFMNLGDHLFAITESNFIRQIDPVTLDTKDK 0 0 VDLAKHLPINIMSSHPLVDGEGNVYTFSSSIFNMGRTKYNLLKFPAAAP 1 2 GTPLETILSQSESICSIDSSWRVSPSYHHSFAMSEKYAVFVEMPLKIDIPKMAVAHLRHMCYSDCIEVLEDTK 0 0 TRIYLVNKETGKQHPITFLCDPLIVYHHVNAYDDGDHVVLDLSCYKKNSFYDKFTMSNLEKTPQEFSKLFDSDEQAVKAMRIVLPLANDS 0 0 KTTGNLVSVANTSCTAEFQGNNIFCTSEMLSVGTECAVINNKYIGKKYKYFYSPGGLKLPPGEM 0 0 LTKIDVETKQRVQTWQEKGCWASQPVFVAKPGATQEDE 1 2 GILMSSVVNENGNPFLLMLDAKSFTEVARIHFDANIPPDVHGVFVPKA* 0
Regularized reference gene collection: teleosts
It is easy to compile large sets of bony vertebrate exons for these three genes using the 44-species genomic alignments at UCSC. To do this, look up the human gene by name at GeneSorter (or blat in a reference sequence) and click on "protein fasta" on the gene details page. Among the various output configurations provided, select all species checking the options as below. Note these alignments, being exon-aware, are intrinsically homological unlike blast alignments in gappy regions which lack this local constraint.
[[Image:]]
For some species, no data is available (shown as dashes). For others, some exons or parts of exons are misssing. This can be due to incompleteness of the respective genome or technical difficulties at exon edges due to split codons. To facilitate uniform comparison of paralogs, the output can be 'regularized' by filling in missing data using the nearest species in the taxonomic sense, for example using chicken data if finch is missing an exon. Most human genes can be completely regularized back to teleost fish. In some cases, notably frog, it may be necessary to stub in an orthologous region from salamander cDNA.
This introduces artefacts that have the effect of understating comparative genomics variability at the locus but that error is offset by the benefits of having 'complete' copies of the three genes in the same set of 44 species. As long as regularization is limited (topologically) to species diverging off the same node (eg lizard could regularize finch and chicken but not frog or platypus), the impact on gene history at the ancestral nodes leading to human will be very minimal. The regularized set works quite well in a blast classificatory tool.
The sequences for RPE65, BCMO1, and BCO2 have been completely regularized in this manner below. (Regularization can be done by hand in a spreadsheet using fill-down and similar commands.) This allows comparison of variation by various techniques such as percent identity at nodes (ie averaging human matches to all species coming off that node if ancestral sequences are not computed) and residue-by-residue conservation by Multalin and similar tools.