Opsin evolution: ancestral introns: Difference between revisions
Tomemerald (talk | contribs) |
Tomemerald (talk | contribs) |
||
| Line 183: | Line 183: | ||
Intron interpretation is further affected by the phenomenon of intron loss followed by later re-intronation. The loss could arise from retropositioning, a process that sweeps out old introns in creating a new genetic locus, or from recombination with a gene's processed mRNA, perhaps purging some 3' introns. Zebrafish and drosophila genomes such retroprocessed opsins. To the extent later intron gain occurs with random position and phase, this results in partial or even full discordancy among paralogous or even orthologous loci or worse, occasional homoplasy of new introns to old. The slow rate of intron events constrains the issue mostly to pre-Cambrian time intervals -- it especially plagues the classification of GPCR and complicates the search for parental genes to opsins. | Intron interpretation is further affected by the phenomenon of intron loss followed by later re-intronation. The loss could arise from retropositioning, a process that sweeps out old introns in creating a new genetic locus, or from recombination with a gene's processed mRNA, perhaps purging some 3' introns. Zebrafish and drosophila genomes such retroprocessed opsins. To the extent later intron gain occurs with random position and phase, this results in partial or even full discordancy among paralogous or even orthologous loci or worse, occasional homoplasy of new introns to old. The slow rate of intron events constrains the issue mostly to pre-Cambrian time intervals -- it especially plagues the classification of GPCR and complicates the search for parental genes to opsins. | ||
Homoplasy in opsins can be reliably detected after considerations of independently determined gene and species trees. For example, a new intron arising post-tunicate pre-lamprey in the LWS stem can hardly be confused even with a fully homoplasic intron that arose in peropsins in pre-bilatera. While a few situations remain ambiguous, the data below -- which surely represents over a hundred billion years of evolutionary branch length -- can be | Homoplasy in opsins can be reliably detected after considerations of independently determined gene and species trees. For example, a new intron arising post-tunicate pre-lamprey in the LWS stem can hardly be confused even with a fully homoplasic intron that arose in peropsins in pre-bilatera. While a few situations remain ambiguous, the data below -- which surely represents over a hundred billion years of evolutionary branch length -- is not hamstrung by homoplasy but instead can be used to empirically estimate the incidence of accidental homoplasic convergence along the opsin gene tree. It seems likely that the outcome would be broadly applicable to both the overall GPCR gene family and the evolution of the overall metazoan proteome. | ||
<pre> | <pre> | ||
Revision as of 11:33, 12 January 2010
See also: Curated Sequences | Alignment | Informative Indels | Ancestral Sequences | Cytoplasmic face | Update Blog
Introduction to intron analysis
Introns within coding regions of opsin genes can potentially provide an independent (or supplemental) means of organizing known opsins into orthologous families and and classifying new ones with ambigous alignment clustering. This becomes especially important as the universe of opsins expands to include rhabdomeric opsins within deuterostomes, ciliary opsins within protostomes, and novel opsins from cnidarians which are otherwise difficult to place (or even distinguish from rhodopsin superfamily non-opsins and other GPCR).
In most lineages, intron pattern is extremely conserved over great evolutionary distances (eg human to anemone), even when amino acid sequence is not. Changes are classified as rare genetic events (RGEs) and can supplement sequence change in determining gene and species tree topology. Other RGEs relevent to opsins include coding indels (insertion or deletion of amino acids) and gene order rearrangements along a chromosome (synteny).
RGEs are characters that can be used in gene tree analyses and reconstruction of ancestral states. Each type of RGE has its own intrinsic time scale that makes it useful on particular aspects of opsin evolution over commensurate time frames. Intron patterns are extremely conserved, making them useless over mammalian, even vertebrate, time scales (stay the same) but are appropriate over Eumetazoa. Indels too are quite conserved (being constrained by membrane width in transmembrane proteins) so are informative within opsins over shorter intervals (eg Pancrustacea). Gene order is only moderately conserved within Bilatera, more commonly it is completely washed out.
All RGEs are potentially subject to homoplasy -- two or more separate events with the same outcome. However, rare events are seldom fixed. Homoplasy amounts to a low probability squared. With an event rate, say for intron gain in a coding gene from last common ancestor with cnidarian to human, not approaching one per billion years per gene, with the average protein having 450 residues and with introns having 3 possible insertion phases at each residue, homoplasy is a total non-issue for the entire proteome (provided intron gain is random).
Intron loss is more frequent but still rare in most lineages. Here there is greater opportunity for homoplasy (notably in Insecta) because the 3' end of the gene is more susceptible to repeats of the mechanism (apparently recombination with retroprocessed mRNA). More intensive taxon sampling can often distinguish timing of separate events. This requires genome sequencing because mature transcripts have lost all information about introns. Uncommonly transcripts retain introns and pseudogenes contain information about ancestral introns. (However opsins, not being transcribed in the germ line to any extent, rarely give rise to retro pseudogenes.)
The vast majority of introns were created in single-celled eukaryotes in the pre-Cambrian. Modulo intron gain and loss, these have descended unchanged in position and phase to the present day. Intron drift (movement by a few residues) does occur but is greatly over-stated when annotation of homologs is sloppy. Intron positions are randomly sited with respect to protein domains. Falsely stated to occur at domain boundaries, some authors are confused by domain iteration (internal tandem duplication of exons by improper recombination) and by domain shuffling.
The first task in utilizing introns as evolutionary characters is to resolve intron gain from loss. This can only be done up to parsimony because the proposition of modelling mechanistically uncertain processes a billion years back in highly diverged lineages (for maximal parsimony) is preposterous. However, provided evenhanded taxonomic sampling is available, the event history is seldom in doubt (rare events squared).
Consequently, the ancestral intronation can be reliably worked out for almost any protein at each species divergence node. While of some intrinsic interest, the main application is evolution of large gene families. Here paralogous branches can have quite different histories. This allows differentiation of these branches from each other at a time when linear sequence homology might become an uncertain guide.
For example melanopsins and encephalopsins are intronated quite differently, even though ultimately both are descended from a single gene. At the time of Ur-bilateran divergence, the intronation of melanopsins has completely coalesced within protostomes but not quite with deuterostomes and not at all with ancestral intronation of ciliary opsins. Consequently the Ur-bilateran had at least two opsins (ie the opsins of fruitfly and human are only homologous, not orthologous). To date, all cnidarian and ctenophore opsins have been single exons genomically or processed transcripts.
Consequently no informative outgroup exists for bilaterans and ancestral opsin intronation cannot be worked out further. (Nematostella normally retains ancestral exons but apparently not here; intron gain is otherwise too rare past this divergence node to account for bilateran opsin intronation.)
Intron location and phase for dummies
The intron pattern consists of two parameters, location and phase (fractional codon distributed across two exons):
Location is easy to specify homologically in opsins because they contain numerous invariant or near-invariant residues sprinkled along their length that provide multiple internal anchors to alignments. The main potential difficulty occurs near an indel (insertion or deletion). However indels are rarely fixed in the core region of opsins because the transmembrane helices (3.4 residues per turn) do not tolerate disruption of their bundle association geometry or membrane spanning lengths.
Similarly the cytoplasmic face and extracellular loop regions, with the exception of CL3, are too short or too engaged in the conserved interactions of signaling and its regulation. Indels in the amino and carboxy termini, which in many opsin classes are extended and poorly conserved, are a different matter; however exons in these regions tend to be extensions of core exons or narrowly lineage-specific.
It's quite possible for more or less the same intron location to arise repeatedly (convergent evolution), especially when 'same' is slightly muddled by indel ambiguity. However phase determination can often disambiguate the near-proximity issue. Here we must pause to review MolBio 101 because many opsin papers exhibit total unawareness of the phase concept:
Three possibilities exist for intron phase: In phase 00, the splice donor (GT in all known opsins) follows immediately after last triplet codon of an exon and the splice acceptor (AG in all known opsins) immediately precedes the first codon of the next exon. In phase 12, an extra basepair follows the last completed triplet codon and precedes the GT start of the splice donor; two extra base pairs (which fill out the split codon and preserve reading frame) precede the acceptor codon. In phase 21 introns, the overhang is 2 bp at the donor end, balanced by 1 bp overhang at the acceptor, together forming a new 3 bp triplet codon.
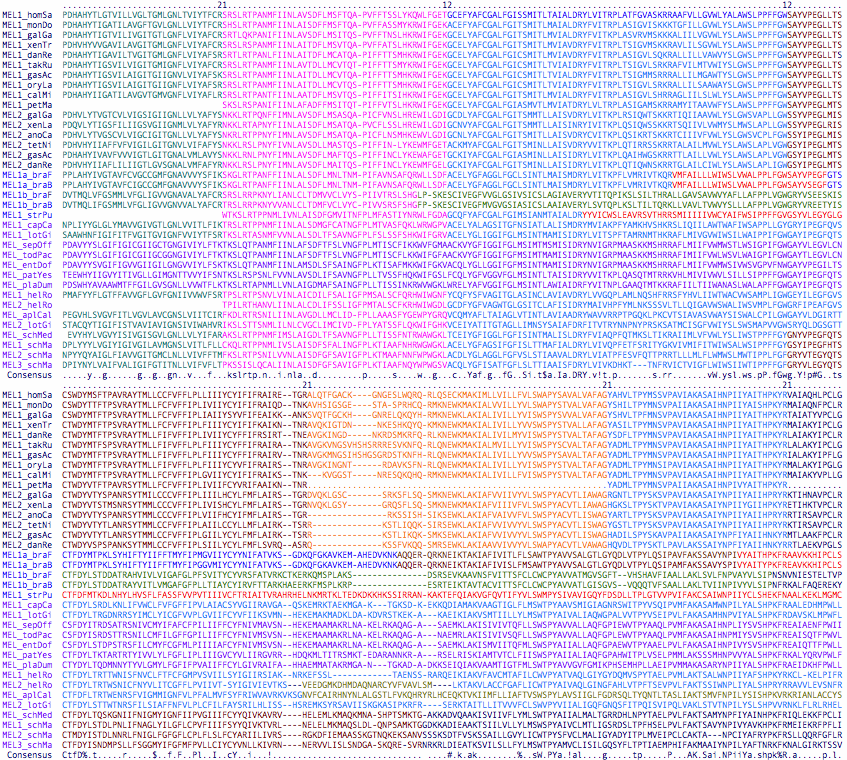
>MEL1_homSap Homo sapiens (human) Gq 483 NM_033282 melanopsin OPN4 0 MNPPSGPRVPPSPTQEPSCMATPAPPSWWDSSQSSISSLGRLPSISPT 0 0 APGTWAAAWVPLPTVDVPDHAHYTLGTVILLVGLTGMLGNLTVIYTFCR 2 1 SRSLRTPANMFIINLAVSDFLMSFTQAPVFFTSSLYKQWLFGET 1 ...
It's useful to indicate phase information within the fasta representation of a sequence. That's done here by line breaks between exons with associated phase overhangs shown by numbers. These numbers are ignored by the vast majority of web software tools so the extra characters do not to be purged before blast queries etc. This format is well-suited to incomplete genome projects because the unit of recovery is typically a whole exon. By convention, the initial methionine is preceded by a 0 even though it is generally part of a larger 5' UTR. Similarily the stop codon asterisk is followed by a 0 even though it is almost always part of a longer 3' UTR. One last convention: the 'extra' amino acid formed by 12 or 21 introns is assigned to the 2 side of the exon break. It's often given incorrectly in Blast output because that tool is not aware of exon breaks and often extends alignments past them into a translated intron.
Normally, phase is determined by aligning by blastn of a transcript (processed already by the cell to remove introns) against genomic sequence. If genomic is not available, the transcript can be reliably intronated in most instances by comparison to a phylogenetically close orthologous gene from a genomic species.
For example, full length transcripts are available for various opsins from the amphioxus Branchiostoma belcheri. However the genome project is not there but over in Branchiostoma floridae. So the B. belcheri proteins need to be placed within the B. floridae assembly, which is conveniently done using Blat on the UCSC genome browser. Exon boundaries are then read off from the alignment details page using 3-frame translation in a second web browser tab at Expasy to ensure smooth reading frame joins and uBlastx against the opsin collection in a third tab to monitor alignment. This process provides a predictive intronation of the presumptive ortholog.
This won't be accurate if B. belcheri has gained or lost introns since the two species diverged. However, outside of certain rogue species, introns typically have a "half-life" of perhaps 5 billion years of branch length, many multiples of the divergence time here. (They're much more conserved than amino acid sequence.) Consequently the inferred gene model will be correct 99% of the time. However every sequence in the Opsin Classifier that originated in a genome project was independently intronated within that project, never by homology. And some species without genome projects (like Platynereis) have the occasional large genomic contig with an opsin.
In practise, it is easy to make small mistakes in assigning phases to genes, especially when percent identity is remote from the alignment query. That's because GT and AG are common dinucleotides and sometimes multiple options seem viable (preserve reading frame). Of course, there's much more to splice sites than just two dinucleotides so sometimes those additional properties must be used to sort out the possibilities (gene prediction software tools carry sophisticated versions of these rules). Usually only one possibility works consistently across the comparative genomics spectrum within a given class of opsins.
It turns out that in post-lamprey deuterostomes not a single case of intron gain or loss can be documented in any of the 14 opsin gene trees (recall each sequence set is maximally phylogenetically dispersed). Since each tree contains several billions of years of branch length, the overall event rate for opsins in this clade is lower than 1 per 50 billion years of evolutionary clock time. Intron conservation of opsins is not atypical -- intron gain and loss is know to be very infrequent (but not zero) across the entire vertebrate proteome.
However other issues must be considered such as alternative splicing, intron sliding, NAGNAG ambiguity, asymmetric frequenciy ratios of phase types (00 is most abundant), mechanisms and relative frequency of gains and loses, hotspots of predisposition, likelihood of convergent evolution, migration out of GT-AG to minor splice forms and so forth. Alternative splicing is irrelevent to opsins because their membrane transiting properties are intolerant -- alternative transcripts are presumably just transcriptome noise. Intron sliding does occurs rarely but most literature claims for it have been debunked. Acceptor ambiguity is real but not seen yet in opsins where indels are disfavored.
There is a general predisposition to enhanced intron loss distally (3') due to recombination with processed mRNA; mechanisms of intron gain are a bit cryptic and -- like everything else -- cannot be assumed the same across all of Metazoa. General trends need not be applicable to the opsing gene family,. Homoplasy hotspots may have some relevance to insect opsins; these apply to inaccessible ancestral sequence so their basis cannot be inferred than contemporary forms.
Using the full inventory of metazoan genome projects, over 350 phylogenetically dispersed opsins in all opsin families have been intronated using direct genomic comparison when possible and homological annotation transfer when not. The error rate is not zero; anomalies needing revisiting are concentrated in 12 and 21 introns and in opsins without close homologs. When a gene model is fragmentary, only half the splice site may be available so that validation of the other half is lacking. In some assemblies, there seem to be sequencing errors that don't allow introns where they are required to avoid premature stop codons.
The scientific literature on intron antiquity is hopelessly muddled due to wild speculation in the pre-informatics era. Today we know that the vast majority of human introns are very old and stable, dating back to early unicellular eukaryotes (eg introns in human SUMF1 are shared with diatom). Consequently most exons were already well entrenched at the time of Eumetazoan emergence and experienced little gain or loss outside of rogue lineages (notably sea urchins, tunicates, nematodes, fruitflies). We expect most deuterostome introns to be present in at least some species of ecdysozoa, lophotrochozoa, and cnidaria; this has been validated recently in the case of Nematostella.
The practical consequence for opsins is that most-- but not all intronation -- occured prior to the major gene duplications and subsequent divergence. To the extent this is valid, a core set of intron location and phases will be common to all opsins. After these are removed, the remaining later-created introns can sometimes guide the reconstruction of the gene tree (independently of sequence alignment). That is, if a series of gene duplications takes place over time, a series of one-off intron creations during the same timeframe will affect only the descendent sub-clade.
Here we expect melanopsins and cilopsins to be distinuished by shared introns from peropsins, neuropsin and RGRopsins. The latter group of opsins, due to their highly diverged nature, have never been persuasively assigned a position in the overall opsin gene tree. However this endeavor requires an extensive collection of cnidarian and earlier branching genomes. To date, opsin candidates in these species have either been intronless (presumably retroprocessed genes like olfactory GPCR) or have not had determinable introns (arose as transcripts).
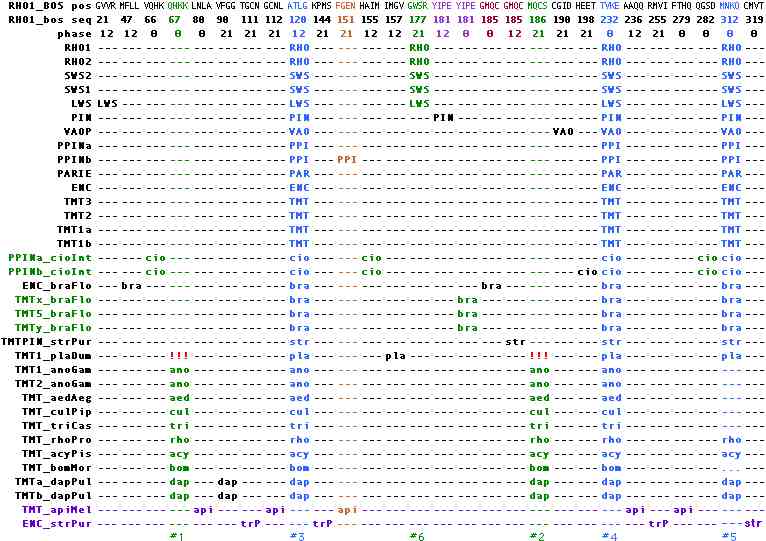
Ancestral ciliary opsin intronation
The ancestral intron pattern in ciliary opsins can be unambiguously determined on the Urbilatera stem as shown below. The exon structure of ciliary opsins no doubt goes back much further in Metazoa but no introns have been described in pre-Bilatera as of Jan 2010. That's unfortunate because at some point ciliary opsin gene structure must coalesce with that of melanopsins, peropsins and ultimately a parental GPCR gene.
Commonsense parsimony is more appropriate than statistical approaches because these simply bury their subjectivity within rarely discussed model assumptions that lack empirical support across the vast time and divergence scales involved here. Predictions about ancestral introns are easily tested by further sequencing in ctenophores and cnidaria.
Intron phases -- important to differentiating introns agreeing in sequence position -- are explained above. Two detailed examples in Annotation Tricks section explain how the Opsin Classifier sequence collection can be used in conjunction with uBlast to determine whether exon breaks of a given opsin agree with another. Gappy regions can require careful curational alignment.
To procede with the actual work of ancestral intron determination, it's helpful to first reduce the number of sequences to a smaller set of proxy sequences that retain all the information but less of the clutter. Proxy sequences can also carry encode indel and synteny (in their header). Homoplasy rarely occurs with respect to position but those situations are disambiguated in every instance by phase or utter remoteness of coinciding events. No evidences supports positional drift, phase change or predisposition to intronation, all dubious propositions to begin with. Introns are conveniently described relative to their position in bovine rhodopsin (which fortuitously exhibits the ancestral pattern).
Sequences can be optimized in various ways to allow more reliable homological comparisons of intron positions to other opsins, including remotely related proteins. Options include ancestral reconstruction, consensus sequence, profile sequence, basal diverging species sequence (lamprey), or a single-species-consistent set (frog would work). However accuracy of ancestral sequences is not experimentally validatable -- reconstruction errors arise in co-evolving but non-adjacent amino acids. Here, actual amniote representative sequences were taken from high quality assemblies.
Within vertebrates, a single proxy sequence suffices to represent each of the 18 distinct genetic loci because intronation patterns change very slowly. It quickly emerges that intronation patterns of ciliary opsins were completely fixed during the tunicate-lamprey stem and have been stable ever since in all lineages (other than in rare retroprocessed genes).
The introns common to all vertebrate ciliary opsins occur at positions 120, 232, and 312 in bovine rhodopsin numbering with phases 12, 00, and 00 and are accurately locatable in alignments using ATLG, TVKE, and MNKQ as text search tags. These can be adequately represented in position-phase notation as 120-12, 232-00, and 312-00. These introns also occur unambiguously in tunicates, amphioxus, sea urchin, insects, crustacean and ragworm.
Here the known gene tree structure readily distinguishes intron gain from intron loss: LWS experienced a gain of 21-12 because all other ingroup and outgroup sequnces have lack this intron. Similarly pinopsin acquired an new intron 181-12, as did VAOP at 190-21. Each intron gain affected a single genetic locus in all species from lamprey on, meaning the loci had already differentiated. This contrasts with the intron gain that occured at 177-21, affecting LWS and all its descendent genes. Note the gain of LWS of 21-12 in LWS must postdate the gain at 177-21 because SWS2 etc were not affected by it.
This implies, contrary to received wisdom, that intron gain was vastly more frequent in the post-tunicate, pre-lamprey ancestor than in the succeeding 500 myr where not a single event occured over many billions of years of branch length. In most genes, intron losses exceed intron gains by a wide margin but this transitional era for vertebrates may be exceptional. The exact sequence of events may never be resolved because of an insufficient number of extant species to sample the tunicate-lamprey stem. Here hagfish offer the most exciting possibility, though no work on them is underway.
The situation is more complex in non-vertebrate deuterostomes, in part because of limited taxonomic sampling but also because of intron churning in fast evolving lineages. Eight idiosyncratic gains but not a single loss are evident in tunicate, amphioxus and sea urchin. (Acornworms lost all ciliary opsins.) Among the 220 intronated ciliary opsins in the curated reference collection, only one sea urchin opsin (very divergent but with key residues intact) has an utterly inexplicable intronation pattern. Perhaps it is misclassified as ciliary.
The intron data in ciliary opsins greatly constrains timing of speculative scenarios of 1R or 2R whole genome duplication in pre-lamprey deuterostomes. (Recall here that amphioxus, despite missing out on these duplications, somehow has *more* genes than human!) That cannot have played any role in any post-encephalopsin, post-amphioxus ciliary opsins which instead are simply sequentially nested intron-preserving segmental duplications with many one-off non-replicated events. The data also conflict with sweeping theories of ectopic propagation of established visual systems via blocks of gene duplication and neofunctionalization.
Although the vast majority of known ciliary opsins reside in deuterostomes, those in protostomes ultimately arbitrate the ancestral intron determination because they constitute the outgroup until relevent sponge, ctenophore and cnidarian opsins are intronated. In ecdysozoa, ciliary opsins are available from crustaceans and certain insects but have been completely lost in lineages such as Drosophila. These opsins also contain the 3 ancestral deuterostome introns (though 312-00 has been lost in mosquitoes and beetle).
Two additional ancestral intron candidates, at 67-00 and 186-21, occur in all ecdysozoan ciliary opsins but are lacking in the sole intronatable lophotrochozoan ciliary opsin (indicated by !!! in the figure) as well as all deuterostomes. Since the currently prefered tree calls for ((edysozoa, lophotrochozoa),deuterostomes), two independent loss events would be required to make these introns ancestral. It is more parsimonious to attribute 67-00 and 186-21 to intron gain in the insect + crustacean stem. This sit
In Lophotrochozoa, the situation is limited to just Platynereis ciliary opsins. Perhaps with targeted sequencing effort, new cdna, additional bioinformatics, or more complete genomes, homologs will emerge in Capitella, Helobdella, Aplysia, Lottia, Schmidtea, or Schistosoma but it appears that ciliary opsins are severely depleted in this large clade. These species provide intronated opsins of melanopsin amd peropsin class more consistently.
In Ecdysozoa, ciliary opsins can be ruled out in the (truly finished) Drosophila genome. The list of species with a ciliary opsin (presumably a single orthologous locus) can be readily extended to Culex, Aedes, Tribolium, Bombyx, Rhodnius, Acyrthosiphon, Heliothis and Daphnia (the only non-insect to date with ciliary opsins). However gene loss seems to have happened repeatedly (or current genomic coverage is insufficient); ciliary opsins cannot be located in Nasonia, Ixodes, and other species with completed genome projects. Nematodes have no opsins of any kind.
Ancestral melanopsin intronation
The well-behaved melanopsin class of opsins was initially defined by an index sequence recovered from frog lateral melanophores in 1998 and further studied in eye and pineal. Its novel role in dispersing light-adsorbing pigment cells raises interesting issues about 'ectopic' expression and the versatility of opsin signaling.
Propagating out from this index sequence to its orthologs and gene duplicates in earlier diverging species ultimately defines a large gene tree encompassing invertebrate opsins while still excluding cilopsins and peropsins. The set of 36 includes 15 from lophotrochozoa of which 8 are intronatable. While blast clustering and alignment-based gene trees continue to be effective, at larger evolutionary distances, opsin relationships become obscured as percent identity approaches the 'floor' of 25% with miscellaneous non-opsin GPCR. Here introns, which evolve much more slowly than most opsin amino acid positions, offer an independent tool for resolving ancient relationships.
The data situation for melanopsins is more favorable in lophotrochozoa and ecdysozoa than for the other two classes. Data distribution is a bit lopsided with 'too many' sequences determined in insects (often non-genomic species) and none in the earliest diverging arthropod branches that are more informative for comparative genomics. It is feasible to compensate for rapid sequence divergence by using greater densities of proxy sequences with known intronation.
It quickly emerges that frog melanopsin and its expansion class within deuterostomes have five nearly perfectly invariant introns that are completely disjoint in position and phase from cilopsins and peropsins. This conservation extends to exactly the same subset of lophotrochozoa opsins independently classified as melanopsins by blast alignment clustering. Thus these opsins do not require a separate name but should simply be denoted as melanopsins to reflect their intronation-defined orthology.
Further, it also emerges that all ultraviolet ecdysozoan opsins (but no others) have these same five conserved introns, though not every opsin in every species conserve them as intron turnover has been much more rapid in panarthropod lineages than in vertebrates. Thus these opsins should simply be called ultraviolet melanopsins to reflect their unequivocal intronation-defined orthology to the genetic locus defined by frog melanopsin. This classification too is fully compatible with that based on sequence similarities.
Ecdysozoa contain another quite distinct class of longer wavelength opsins ('kumopsins') that cluster to melanopsins much more closely than to cilopsins or peropsins, yet have a totally distinct pattern of 5-6 other conserved introns that must reflect a much older divergence. In Drosophila, these have been denoted historically as Rh1, RH2 and Rh6. The phylogenetic distribution of this opsin class encompasses hexapods and chelicerates but no counterpart has survived in living lophotrochozoa or deuterostomes. It too must have been present in the ur-bilateran as a second melanopsin, presumably still adapted to longer wavelengths.
The ur-bilateran evidently possessed a melanopsin containing these five introns. Since melanopsins have not yet coalesced with peropsins or cilopsins in cnidaria and ctenophores, it can be safely predicted that these introns will eventually be found in opsins of these early metazoa (which are known to be quite conservative overall in intron retention. This ur-bilateran also hosted at least one ciliary opsin and at least three peropsin-class opsins, again based either solely on intronation patterns or on sequence coalescence. Any common ground to these gene classes is far more deeply ancestral.
Ecdysozoa contain a final group of related opsins known solely from ten crustacean and one chelicerate sequence. These are currently unclassifiable by intronic criteria because they are known strictly from processed transcripts, other than one gene from the intron-churning species Daphnia. While quite diverged and possessing various idiosyncratic indels and diagnostic residues, they seem to classify most closely with the long wavelength melanopsins. Introns here could help determine whether or not they deserve separate status. Early diverging arthropods may contain yet other classes of ancient opsins.
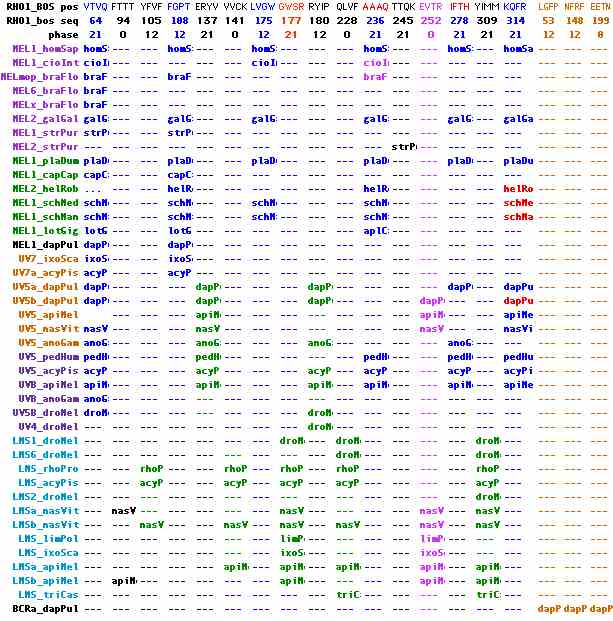
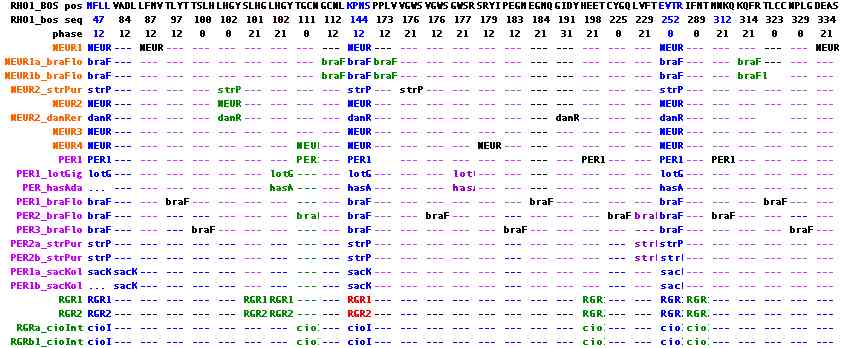
Ancestral peropsin, neuropsin and RGRopsin intronation
Peropsins, rgropsins and neuropsins are commonly taken as a self-contained subgroup in terms of both sequence clustering and their set of unique introns, though exactly how they are nested within the topology of other opsin classes is not completely clear. Exon breaks are colored in the first accompaning image with phases shown in the top line. Four molluscan peropsins to serve as outgroup to the otherwise entirely deuterostomic collection, proving their presence in Urbilatera (which was already apparent from deep rooting).
The first exon break of phase 12 is shared by all 35 members and hence is ancestral. A long second exon, shown in red and also ending in phase 12, is also universally shared distally (though in vertebrates shortened by 3 residues). In all but 3 deeply diverging peropsins, it is broken into two exons in 6 different ways utilizing the 3 different phases. A third universal exon break occurs near the end of the protein. It too can have various internal introns.
These sporadic introns follow within-class blastp cluster associations, though some shared endpoints suggest alternative scenarios. It's important to realize most parsimonius scenario is not necessarily the actual history -- which is a one-off sequence of events for any given gene family, not a statistical ensemble. It would be especially helpful to locate intronated cnidarian opsins in this group.
The second figure includes two new ecdysozoan peropsins, presenting the data in position-phase notation relative to bovine rhodopsin. This spreadsheet visualization will eventually allow intron comparisons among all opsins. It can be seen that three ancestral introns are shared among all peropsins, rgropsins and neuropsins, namely 47-13, 144-12, and 252-00. These are completely distinct from the three ancestral ciliary opsins and no plausible amount of 'intron sliding' can inter-relate them. There is no support for the two candidate ancestral ciliary introns seen only in ecdysozoa.
Note further that the lophotrochozoan peropsins share two sporadic introns (102-21 and 177-21) with the one intronatable ecdysozoan peropsin (in addition to the universal introns). This strongly supports the standard topology of bilaterans in regards to deuterostomes. An sporadic intron of NEUR1_strPur at 102-00 shared with NEUR2 opsins, along with the lack of two introns found in NEUR1, suggests the sea urchin opsin need reclassification to the NEUR2 group but the ur-neuropsin within deuterostomes remains unresolved because of the amphioxus situation. Indeed it appears that the large number of intron gains has resulted in some homoplasy.
Gapping ambiguity can be a serious issue when introns happen to fall in non-transmembrane loops where length is not necessarily well constrained; some regions lack satisfactorily conserved biflanking anchoring residues. However in the case of the three universal introns, this can be turned around to significantly constrain gapping. This has applications in RGR at 144-12 where software not embodying the intron constraint will mis-gap the alignment.
Neuropsins have a two residule indel in the EC2 loop region with reliable conserved flanking CTLDWWLAQASVGGQVF; that length is seen again in cone and rod opsins. Therefore the indel is an insertion event and does not serve to unite peropsins and rgropsins. The same can be said for a 3 residue deletion in RGR beginning at position 143 (again bovine rhodopsin coordinates: indel notation gives start coordinate, length, and resolution so 143:-3 here).
In summary, neither introns nor indels clarify the proper grouping of peropsins, rgropsins and neuropsins with respect to each other. However the introns do provide overwhelming evidence that these three opsin families must cluster together (ie apart from melanopsins and cilopsins), deriving from a single coalesced ancestral gene with these ancient introns. Because the primary intronation era was so far back in time, it may well be that peropsins, rgropsins and neuropsins originated from a different parental GPCR gene than melanopsins and cilopsins.
Ancestral introns
We're now in a far better position to analyze putative opsins in cnidaria, ctenophores and sponges which might be seriously diverged in primary sequence. Nematostella in particular is quite conservative overall in terms of ancestral intron retention (minimal gain and loss relative to human). Opsins so far have been an exceptional gene family in that regard because all the K296 GPCR to date appear to have arisen from retropositioning (as in olfactory genes).
A weak blast match to authentic opsins and proven expression in a photoreception cell is insufficient to establish a given candidate gene as an opsin: a slow-evolving generic GPCR might also give similar alignment quality and many other signaling processes take place simultaneously even within specialized cell types. Without diagnostic residues, appropriate introns, and informative indels, the evidence could be very circumstantial. In fact, there may be ciliary pre-opsins within the rhodopsin GPCR superfamily which are not engaged in photoreception themselves but survive as members of the immediate sister gene family. That could account for the excessive numbers being reported in cnidaria vis-a-vis their visual requirements and also their independent intronation.
More provocatively, certain non-opsin GPCR may still contain these introns. If these have leading blast scores, an intriguing case could be made for these being the long-sought parental GPCR to opsins. Unfortunately many of those in the GPCR outgroup reference collection are intronless or have a single intron that may reflect re-intronation after a retroprocessing sweep. However it is useful to pull together the major conserved introns from cilopsins, peropsins and melanopsins in order that the introns can be compared over this vast range of sequences.
Managing homoplasy in intron data
Coding region intron data in comma-delimited format: paste into a spreadsheet for more convenient analysis. Most of the rows are proxies where a single sequence (eg bovine rhodopsin) represents dozens if not hundreds of identically intronated orthologs. Every sequence in the curated reference collection with genomically determinable intronation is present as itself or as proxy. Early and late introns are not included if outside the alignable region (which barely extends beyond the heptatransmembrane core) because introns cannot be assigned reliable coordinates relative to RHO1 when gapping uncertainties are overwhelming.
Columns with only 1-2 entries generally represent sporadic introns limited to a narrow clade or tandem gene duplicate pair. Certain species such as drosophila, sea urchin, amphioxus and tunicate have relatively high rates of intron gain and loss, resulting in unique introns. These columns can be considered noise in terms of gene family phylogenetic analysis and deleted from the spreadsheet to simplify it. However they have some value in providing homoplasy statistics.
Homoplasy may be positional (accidental agreement in position with or without agreement in phase). While not common, roughly a third of these will coincidentally match in phase (full homoplasy). A typical opsin might have 350 residues of which 300 core residues are broadly alignable. Given 3 phases and so 900 possible intronations, full homoplasy would seem statistically implausible.
Yet genomewide, phases are used disproportionately (roughly half are 00), lowering the capacity of phase to resolve positional homoplasy. Recalling from high school mathematics that a room with 57 people, the probability is surprisingly 99% that two have the same birthday despite 365 days in the year. Thus probabilities for full homoplasy in opsin introns, calculated as collisions in a hash table, are unexpectedly high.
This lead to a foolish literature asserting genes have structural or compositional hotspots predisposing them to repeat intronation. These analyses invariably fail to consider that intronation happened in inaccessible ancestral rather than contemporary sequences. Within opsins, the discussion has centered on the GWSR region (position 177 in bovine rhodopsin) which exhibits both positional and full homoplasy. However other instances can be located below.
However it is hazardous to compute a priori likelihoods because actual historical mechanisms for gain and loss of specific introns remain murky. It has not been established that observable contempory mechanisms are the same as in the past. Partial recombination with retro-processed mRNA may explain some intron loss; recombination across flanking retroposons has also been invoked. It is abundandly clear that, with the exception of internal tandemly repeated domains in protein concatenates, neither intron position nor phase correlate meaningfully with amino acid or base composition, pfam domains, folding units, secondary structure, transmembrane helices etc.
Opsin genes are pecularly short (ie their introns contain few retroposons), with unknown implications for their intron history. That is, the median span of a comparably sized human coding gene might be 100 kbp whereas RHO1 is 5% of that length. Short introns, say in genes encoding ribosomal proteins, are sometimes associated with high rates of transcription (ie providing energetic savings to retroposon purging) but this seems inapplicable to opsins which can be quite rare or even non-existent in massive whole-animal cDNA collections.
For all intents and purposes, introns appear to be inserted randomly in position, without any selectional consequences other than minimal size constraints on spliceable exons. Most proteins, like opsins, seem to have acquired their introns very early on, an era followed by preservation for tens of billions of years of branch length (though not total stasis in all lineages). This makes intron signatures very useful tools for gene tree phylogeny, provided inevitable instances of homoplasy can be 'managed'.
Opsin introns did not arise evenly over geologic time (ie from a temporally uniform stochastic process). For example, intron gain was quite common in stem deuterostomes but all but ceased after the Cambrian in vertebrates. Similarly drosophilids and nematodes churned introns at such rates that phylogenetically more remote pre-bilaterans have a much better overall intron match with humans. For modeling purposes, separate probablility distributions are needed on internode stems. Here fixation of intron events within an ancestral population must be distinguished from rates of events which were not fixed and are not ancestrally observable.
Intron interpretation is further affected by the phenomenon of intron loss followed by later re-intronation. The loss could arise from retropositioning, a process that sweeps out old introns in creating a new genetic locus, or from recombination with a gene's processed mRNA, perhaps purging some 3' introns. Zebrafish and drosophila genomes such retroprocessed opsins. To the extent later intron gain occurs with random position and phase, this results in partial or even full discordancy among paralogous or even orthologous loci or worse, occasional homoplasy of new introns to old. The slow rate of intron events constrains the issue mostly to pre-Cambrian time intervals -- it especially plagues the classification of GPCR and complicates the search for parental genes to opsins.
Homoplasy in opsins can be reliably detected after considerations of independently determined gene and species trees. For example, a new intron arising post-tunicate pre-lamprey in the LWS stem can hardly be confused even with a fully homoplasic intron that arose in peropsins in pre-bilatera. While a few situations remain ambiguous, the data below -- which surely represents over a hundred billion years of evolutionary branch length -- is not hamstrung by homoplasy but instead can be used to empirically estimate the incidence of accidental homoplasic convergence along the opsin gene tree. It seems likely that the outcome would be broadly applicable to both the overall GPCR gene family and the evolution of the overall metazoan proteome.
RHO1_BOS pos,GVVR,MFLL,VQHK,QHKK,LNLA,VFGG,TGCN,GCNL,ATLG,KPMS,FGEN,HAIM,IMGV,GWSR,YIPE,YIPE,GMQC,GMQC,MQCS,CGID,HEET,TVKE,AAQQ,RMVI,FTHQ,QGSD,MNKQ,CMVT,,,,, RHO1_bos seq,21,47,66,67,80,90,111,112,120,144,151,155,157,177,181,181,185,185,186,190,198,232,236,255,279,282,312,319,,,,, phase,12,12,0,0,0,21,21,21,12,21,21,12,12,21,12,0,0,12,21,21,21,0,12,21,0,0,0,0,,,,, RHO1,---,---,---,---,---,---,---,---,RHO1,---,---,---,---,RHO1,---,---,---,---,---,---,---,RHO1,---,---,---,---,RHO1,---,,,,, RHO2,---,---,---,---,---,---,---,---,RHO2,---,---,---,---,RHO2,---,---,---,---,---,---,---,RHO2,---,---,---,---,RHO2,---,,,,, SWS2,---,---,---,---,---,---,---,---,SWS2,---,---,---,---,SWS2,---,---,---,---,---,---,---,SWS2,---,---,---,---,SWS2,---,,,,, SWS1,---,---,---,---,---,---,---,---,SWS1,---,---,---,---,SWS1,---,---,---,---,---,---,---,SWS1,---,---,---,---,SWS1,---,,,,, LWS,LWS,---,---,---,---,---,---,---,LWS,---,---,---,---,LWS,---,---,---,---,---,---,---,LWS,---,---,---,---,LWS,---,,,,, PIN,---,---,---,---,---,---,---,---,PIN,---,---,---,---,---,PIN,---,---,---,---,---,---,PIN,---,---,---,---,PIN,---,,,,, VAOP,---,---,---,---,---,---,---,---,VAOP,---,---,---,---,---,---,---,---,---,---,VAOP,---,VAOP,---,---,---,---,VAOP,---,,,,, PPINa,---,---,---,---,---,---,---,---,PPINa,---,---,---,---,---,---,---,---,---,---,---,---,PPINa,---,---,---,---,PPINa,---,,,,, PPINb,---,---,---,---,---,---,---,---,PPINb,---,PPINb,---,---,---,---,---,---,---,---,---,---,PPINb,---,---,---,---,PPINb,---,,,,, PARIE,---,---,---,---,---,---,---,---,PARIE,---,---,---,---,---,---,---,---,---,---,---,---,PARIE,---,---,---,---,PARIE,---,,,,, ENC,---,---,---,---,---,---,---,---,ENC,---,---,---,---,---,---,---,---,---,---,---,---,ENC,---,---,---,---,ENC,---,,,,, TMT3,---,---,---,---,---,---,---,---,TMT3,---,---,---,---,---,---,---,---,---,---,---,---,TMT3,---,---,---,---,TMT3,---,,,,, TMT2,---,---,---,---,---,---,---,---,TMT2,---,---,---,---,---,---,---,---,---,---,---,---,TMT2,---,---,---,---,TMT2,---,,,,, TMT1a,---,---,---,---,---,---,---,---,TMT1a,---,---,---,---,---,---,---,---,---,---,---,---,TMT1a,---,---,---,---,TMT1a,---,,,,, TMT1b,---,---,---,---,---,---,---,---,TMT1b,---,---,---,---,---,---,---,---,---,---,---,---,TMT1b,---,---,---,---,TMT1b,---,,,,, PPINa_cioInt,---,---,cioIn,---,---,---,---,---,cioIn,---,---,cioIn,---,---,---,---,---,---,---,---,---,cioIn,---,---,---,cioIn,cioIn,---,,,,, PPINb_cioInt,---,---,cioIn,---,---,---,---,---,cioIn,---,---,cioIn,---,---,---,---,---,---,---,---,cioIn,cioIn,---,---,---,cioIn,cioIn,---,,,,, ENC_braFlo,---,braFl,---,---,---,---,---,---,braFl,---,---,---,---,---,---,---,braFl,---,---,---,---,braFl,---,---,---,---,braFl,---,,,,, TMTx_braFlo,---,---,---,---,---,---,---,---,braFl,---,---,---,---,---,---,braFl,---,---,---,---,---,braFl,---,---,---,---,braFl,---,,,,, TMT5_braFlo,---,---,---,---,---,---,---,---,braFl,---,---,---,---,---,---,braFl,---,---,---,---,---,braFl,---,---,---,---,braFl,---,,,,, TMTy_braFlo,---,---,---,---,---,---,---,---,braFl,---,---,---,---,---,---,braFl,---,---,---,---,---,braFl,---,---,---,---,braFl,---,,,,, TMTPIN_strPur,---,---,---,---,---,---,---,---,strPu,---,---,---,---,---,---,---,---,strPu,---,---,---,strPu,---,---,---,---,strPu,---,,,,, TMT1_plaDum,---,---,---,!!!,---,---,---,---,plaDu,---,---,---,plaDu,---,---,---,---,---,!!!,---,---,plaDu,---,---,---,---,plaDu,---,,,,, TMT1_anoGam,---,---,---,anoGa,---,---,---,---,anoGa,---,---,---,---,---,---,---,---,---,anoGa,---,---,anoGa,---,---,---,---,---,---,,,,, TMT2_anoGam,---,---,---,anoGa,---,---,---,---,anoGa,---,---,---,---,---,---,---,---,---,anoGa,---,---,anoGa,---,---,---,---,---,---,,,,, TMT_aedAeg,---,---,---,aedAe,---,---,---,---,aedAe,---,---,---,---,---,---,---,---,---,aedAe,---,---,aedAe,---,---,---,---,---,---,,,,, TMT_culPip,---,---,---,culPi,---,---,---,---,culPi,---,---,---,---,---,---,---,---,---,culPi,---,---,culPi,---,---,---,---,---,---,,,,, TMT_triCas,---,---,---,triCa,---,---,---,---,triCa,---,---,---,---,---,---,---,---,---,triCa,---,---,triCa,---,---,---,---,---,---,,,,, TMT_rhoPro,---,---,---,rhoPr,---,---,---,---,rhoPr,---,---,---,---,---,---,---,---,---,rhoPr,---,---,rhoPr,---,---,---,---,rhoPr,---,,,,, TMT_acyPis,---,---,---,acyPi,---,---,---,---,acyPi,---,---,---,---,---,---,---,---,---,acyPi,---,---,acyPi,---,---,---,---,acyPi,---,,,,, TMT_bomMor,---,---,---,bomMo,---,---,---,---,bomMo,---,---,---,---,---,---,---,---,---,bomMo,---,---,bomMo,---,---,---,---,...,---,,,,, TMTa_dapPul,---,---,---,dapPu,---,dapPu,---,---,dapPu,---,---,---,---,---,---,---,---,---,dapPu,---,---,dapPu,---,---,---,---,dapPu,---,,,,, TMTb_dapPul,---,---,---,dapPu,---,dapPu,---,---,dapPu,---,---,---,---,---,---,---,---,---,dapPu,---,---,dapPu,---,---,---,---,dapPu,---,,,,, TMT_apiMel,---,---,---,---,apiMe,---,---,apiMe,---,---,apiMe,---,---,---,---,---,---,---,---,---,---,---,apiMe,---,apiMe,---,---,---,,,,, ENC_strPur,---,---,---,---,---,---,strPu,---,---,strPu,---,---,---,---,---,---,---,---,---,---,---,---,---,strPu,---,---,---,str,,,,, RHO1_BOS pos,MFLL,VADL,LFMV,TLYT,TSLH,SLHG,LHGY,LHGY,TGCN,GCNL,KPMS,PPLV,VGWS,VGWS,GWSR,SRYI,PEGM,EGMQ,GIDY,HEET,CYGQ,LVFT,EVTR,IFMT,MNKQ,KQFR,TLCC,NPLG,DEAS,,,, RHO1_bos seq,47,84,87,97,100,101,102,102,111,112,144,173,176,176,177,179,183,184,191,198,225,229,252,289,312,314,323,329,334,,,, phase,12,12,12,12,0,21,0,21,0,12,12,12,21,12,21,12,12,21,31,21,0,21,0,0,21,21,0,0,21,,,, NEUR1,NEUR1,---,NEUR1,---,---,---,---,---,---,---,NEUR1,---,---,---,---,---,---,---,---,---,---,---,NEUR1,---,---,---,---,---,NEUR1, ,,, NEUR1a_braFlo,braFl,---,---,---,---,---,---,---,---,braFl,braFl,braFl,---,---,---,---,---,---,---,---,---,---,braFl,---,---,braFl,---,---,---,,,, NEUR1b_braFlo,braFl,---,---,---,---,---,---,---,---,braFl,braFl,braFl,---,---,---,---,---,---,---,---,---,---,braFl,---,---,braFl,---,---,---,,,, NEUR2_strPur,strPu,---,---,---,---,---,strPu,---,---,---,strPu,---,strPu,---,---,---,---,---,---,---,---,---,strPu,---,---,---,---,---,---,,,, NEUR2,NEUR2,---,---,---,---,---,NEUR2,---,---,---,NEUR2,---,---,---,---,---,---,---,---,---,---,---,NEUR2,---,---,---,---,---,---,,,, NEUR2_danRer,danRe,---,---,---,---,---,danRe,---,---,---,danRe,---,---,---,---,---,---,---,danRe,---,---,---,danRe,---,---,---,---,---,---,,,, NEUR3,NEUR3,---,---,---,---,---,---,---,---,---,NEUR3,---,---,---,---,---,---,---,---,---,---,---,NEUR3,---,---,---,---,---,---,,,, NEUR4,NEUR4,---,---,---,---,---,---,---,NEUR4,---,NEUR4,---,---,---,---,NEUR4,---,---,---,---,---,---,NEUR4,---,---,---,---,---,---,,,, PER1,PER1,---,---,---,---,---,---,---,PER1,---,PER1,,---,---,---,---,---,---,---,PER1,---,---,PER1,---,PER1,---,---,---,---,,,, PER1_lotGig,lotGi,---,---,---,---,---,---,lotGi,---,---,lotGi,---,---,---,lotGi,---,---,---,---,---,---,---,lotGi,---,---,---,---,---,---,,,, PER_hasAda,...,---,---,---,---,---,---,hasAd,---,---,hasAd,---,---,---,hasAd,---,---,---,---,---,---,---,hasAd,---,---,---,---,---,---,,,, PER1_braFlo,braFl,---,---,braFl,---,---,---,---,---,---,braFl,---,---,---,---,---,---,braFl,---,---,---,---,braFl,---,---,---,braFl,---,---,,,, PER2_braFlo,braFl,---,---,---,---,---,---,---,braFl,---,braFl,---,---,braFl,---,---,---,---,---,---,braFl,braFl,braFl,---,braFl,---,---,---,---,,,, PER3_braFlo,braFl,---,---,---,braFl,---,---,---,---,---,braFl,---,---,---,---,---,braFl,---,---,---,---,---,braFl,---,---,---,---,braFl,---,,,, PER2a_strPur,strPu,---,---,---,---,---,---,---,---,---,strPu,---,---,---,---,---,---,---,---,---,---,strPu,strPu,---,---,---,---,---,---,,,, PER2b_strPur,strPu,---,---,---,---,---,---,---,---,---,strPu,---,---,---,---,---,---,---,---,---,---,strPu,strPu,---,---,---,---,---,---,,,, PER1a_sacKol,sacKo,sacKo,---,---,---,---,---,---,---,---,sacKo,---,---,---,---,---,---,---,---,---,---,---,sacKo,---,---,---,---,---,---,,,, PER1b_sacKol,...,sacKo,---,---,---,---,---,---,---,---,sacKo,---,---,---,---,---,---,---,---,---,---,---,sacKo,---,---,---,---,---,---,,,, RGR1,RGR1,---,---,---,---,RGR1,---,RGR1,---,---,RGR1,---,---,---,---,---,---,---,---,RGR1,---,---,RGR1,RGR1,---,---,---,---,---,,,, RGR2,RGR2,---,---,---,---,RGR2,---,RGR2,---,---,RGR2,---,---,---,---,---,---,---,---,RGR2,---,---,RGR2,RGR2,---,---,---,---,---,,,, RGRa_cioInt,cioIn,---,---,---,---,---,---,---,cioIn,---,cioIn,---,---,---,---,---,---,---,---,cioIn,---,---,cioIn,cioIn,---,---,---,---,---,,,, RGRb1_cioInt,cioIn,---,---,---,---,---,---,---,cioIn,---,cioIn,---,---,---,---,---,---,---,---,cioIn,---,---,cioIn,cioIn,---,---,---,---,--- RHO1_BOS pos,VTVQ,FTTT,YFVF,FGPT,ERYV,VVCK,LVGW,GWSR,RYIP,QLVF,AAAQ,TTQK,EVTR,IFTH,YIMM,KQFR,,NFRF,LGFP,EETN,YNPV,VADL,QKAE,PEGM,LWSL,LHGY,FVVH,FGEN,ATLG,AIER RHO1_bos seq,64,94,105,108,137,141,175,177,180,228,236,245,252,278,309,314,,148,53,199,304,84,247,183,128,102,211,151,120,135 phase,21,0,12,12,21,0,12,21,12,0,21,21,0,21,21,21,,12,12,0,12,21,0,12,0,21,0,0,12,21 MEL1_homSap,homSa,---,---,homSa,---,---,homSa,---,---,---,homSa,---,---,homSa,---,homSa,,---,---,---,---,---,---,---,---,---,---,---,---,--- MEL1_cioInt,cioIn,---,---,---,---,---,cioIn,---,---,---,cioIn,---,---,---,---,---,,---,---,---,---,---,---,---,---,---,---,---,---,--- MELmop_braFlo,braFl,---,---,braFl,---,---,---,---,---,---,braFl,---,---,---,---,---,,---,---,---,braFl,---,---,braFl,---,---,---,braFl,---,--- MEL6_braFlo,braFl,---,---,---,---,---,---,---,---,---,---,---,---,---,---,---,,---,---,---,---,---,---,---,---,braFl,---,---,---,--- MELx_braFlo,braFl,---,---,---,---,---,---,---,---,---,---,---,---,---,---,---,,---,---,---,---,---,---,---,braFl,braFl,---,---,---,--- MEL2_galGal,galGa,---,---,galGa,---,---,galGa,---,---,---,galGa,---,---,galGa,---,galGa,,---,---,---,---,---,---,---,---,---,---,---,---,--- MEL1_strPur,strPu,---,---,strPu,---,---,---,---,---,---,---,---,---,---,---,---,,---,---,---,---,---,---,---,---,---,---,---,---,strPu MEL2_strPur,---,---,---,---,---,---,---,---,---,---,---,strPu,---,---,---,---,,---,---,---,---,---,---,---,---,---,---,---,---,--- MEL1_plaDum,plaDu,---,---,plaDu,---,---,plaDu,---,---,---,plaDu,---,---,plaDu,---,plaDu,,---,---,---,---,---,---,---,---,---,---,---,---,--- MEL1_capCap,capCa,---,---,capCa,---,---,---,---,---,---,---,---,---,---,---,---,,---,---,---,---,---,---,---,---,---,---,---,---,--- MEL2_helRob,...,---,---,helRo,---,---,---,---,---,---,helRo,---,---,---,---,helRo,,---,---,---,---,---,helRo,---,---,---,---,---,---,--- MEL1_schMed,schMe,---,---,schMe,---,---,schMe,---,---,---,schMe,---,---,---,---,schMe,,---,---,---,---,---,---,---,---,---,---,---,---,--- MEL1_schMan,schMa,---,---,schMa,---,---,schMa,---,---,---,schMa,---,---,---,---,schMa,,---,---,---,---,---,---,---,---,---,---,---,---,--- MEL1_lotGig,lotGi,---,---,lotGi,---,---,---,---,---,---,aplCa,---,---,---,---,---,,---,---,---,---,---,---,---,---,---,---,---,---,--- MEL1_dapPul,dapPul,---,---,dapPul,---,---,---,---,---,---,---,---,---,---,---,---,,---,---,---,---,---,---,---,---,---,---,---,---,--- UV7_ixoSca,ixoSc,---,---,ixoSc,---,---,---,---,---,---,---,---,---,---,---,---,,---,---,---,---,---,---,---,---,---,---,---,---,--- UV7a_acyPis,acyPi,---,---,acyPi,---,---,---,---,---,---,---,---,---,---,---,---,,---,---,---,---,---,---,---,---,---,---,---,---,--- UV5a_dapPul,dapPu,---,---,---,dapPu,---,---,---,dapPu,---,---,---,---,dapPu,---,dapPu,,---,---,---,---,---,---,---,---,---,---,---,---,--- UV5b_dapPul,dapPu,---,---,---,dapPu,---,---,---,dapPu,---,---,---,dapPu,---,---,dapPu,,---,---,---,---,---,---,---,---,---,---,---,---,--- UV5_apiMel,,---,---,---,apiMe,---,---,---,---,---,---,---,apiMe,---,---,apiMe,,---,---,---,---,---,---,---,---,---,---,---,---,--- UV5_nasVit,nasVi,---,---,---,nasVi,---,---,---,---,---,---,---,nasVi,---,---,nasVi,,---,---,---,---,---,---,---,---,---,---,---,---,--- UV5_anoGam,anoGa,---,---,---,anoGa,---,---,---,anoGa,---,---,---,---,anoGa,---,---,,---,---,---,---,---,---,---,---,---,---,---,anoGa,--- UV5_pedHum,pedHu,---,---,---,pedHu,---,---,---,---,---,pedHu,---,---,pedHu,---,pedHu,,---,---,---,---,---,---,---,---,---,---,---,---,--- UV5_acyPis,acyPi,---,---,---,acyPi,---,---,---,acyPi,---,acyPi,---,---,acyPi,---,acyPi,,---,---,---,---,---,---,---,---,---,---,---,---,--- UVB_apiMel,apiMe,---,---,---,apiMe,---,---,---,apiMe,---,apiMe,---,---,apiMe,---,apiMe,,---,---,---,---,---,---,---,---,---,---,---,---,--- UVB_anoGam,anoGa,---,---,---,---,---,---,---,---,---,---,---,---,---,---,---,,---,---,---,---,---,---,---,---,---,---,---,---,--- UV5B_droMel,droMe,---,---,---,---,---,---,---,droMe,---,---,---,---,---,---,---,,---,---,---,---,---,---,---,---,---,---,---,---,--- UV4_droMel,---,---,---,---,---,---,---,---,droMe,---,---,---,---,---,---,---,,---,---,---,---,---,---,---,---,---,---,---,---,--- LMS1_droMel,---,---,---,---,---,---,---,droMel,---,droMel,---,---,---,---,droMel,---,,---,---,---,---,---,---,---,---,---,---,---,---,--- LMS6_droMel,---,---,---,---,---,---,---,---,---,droMel,---,---,---,---,droMel,---,,---,---,---,---,---,---,---,---,---,---,---,---,--- LMS_rhoPro,,,rhoPr,---,---,rhoPr,,rhoPr,,rhoPr,---,---,---,---,rhoPr,---,,---,---,---,---,,---,,---,,---,,---,--- LMS_acyPis,---,---,acyPis,---,---,acyPis,---,acyPis,---,acyPis,---,---,---,---,acyPis,---,,---,---,---,---,---,---,---,---,---,---,---,---,--- LMS2_droMel,---,---,---,---,---,---,---,---,---,---,---,---,---,---,droMel,---,,---,---,---,---,---,---,---,---,---,---,---,---,--- LMSa_nasVit,---,nasVit,---,---,---,---,---,nasVit,---,---,---,---,nasVit,---,nasVit,---,,---,---,---,---,---,---,---,---,---,---,---,---,--- LMSb_nasVit,---,---,nasVit,---,---,nasVit,---,nasVit,---,nasVit,---,---,nasVit,---,nasVit,---,,---,---,---,---,---,---,---,---,---,---,---,---,--- LMS_limPol,---,---,---,---,---,---,---,limPol,---,---,---,---,limPol,---,---,---,,---,---,---,---,---,---,---,---,---,limPol,---,---,--- LMS_ixoSca,---,---,---,---,---,---,---,ixoSca,---,---,---,---,ixoSca,---,---,---,,---,---,---,---,ixoSca,---,---,---,---,---,---,---,--- LMSa_apiMel,---,---,---,---,---,apiMel,---,apiMel,---,apiMel,---,---,apiMel,---,apiMel,---,,---,---,---,---,,---,---,---,---,---,---,---,--- LMSb_apiMel,---,apiMel,---,---,---,---,---,apiMel,---,---,---,---,apiMel,---,apiMel,---,,---,---,---,---,---,---,---,---,---,---,---,---,--- LMS_triCas,---,---,---,---,---,---,---,---,---,triCas,---,---,---,---,triCas,---,,---,---,---,---,---,---,---,---,---,---,---,---,--- BCRa_dapPul,---,---,---,---,---,---,---,---,---,---,---,---,---,---,---,---,,dapPul,dapPul,dapPul,---,---,---,---,---,---,---,---,---,---
See also: Curated Sequences | Alignment | Informative Indels | Ancestral Sequences | Cytoplasmic face | Update Blog