Making a hub for a cell browser
This page is divided into two sections:
- A discussion of makeCbHub and it's options
- How to make a track hub for a cell browser utilizing makeCbHub
makeCbHub
makeCbHub will generate the trackDb stanzas for a set of tracks that you can then use to create a track hub. This section will go through the various options for the script and how to use them
Required option: fileDir
There is one required option: fileDir. This is a directory of big* files or a directory of directories containing big* files.
For example, use the commands below to generate the trackDb stanzas for a single composite track in the mouse-brain-cutandtag dataset:
cd /hive/data/inside/cells/datasets/mouse-brain-cutandtag/hub makeCbHub olig2
Output:
track olig2
compositeTrack on
shortLabel olig2
longLabel olig2
visibility dense
autoScale group
type bigWig
track olig2_cluster_non_oligo
parent olig2 on
shortLabel cluster_non_oligo
longLabel cluster_non_oligo
type bigWig 0.000000 2358.294189
autoScale group
bigDataUrl olig2/cluster_non_oligo.bw
visibility dense
track olig2_cluster_oligo
parent olig2 on
shortLabel cluster_oligo
longLabel cluster_oligo
type bigWig 0.000000 280.645325
autoScale group
bigDataUrl olig2/cluster_oligo.bw
visibility dense
As you can see, it works, though it wouldn't be particularly pretty to look at the Genome Browser. The labels are not very human-friendly and both tracks will be colored the same, default color, black.
Options to customize output
The six optional arguments for makeCbHub allow you greater control over what’s put into these trackDb stanzas, including shortLabels, longLabels, and colors.
Composite track labels: -d/--datasetList
The option -d/--datasetList serves two purposes:
- If
fileDircontains multiple dirs, you can specify which of those you want to build trackDb stanzas for - By default, the directory names under
fileDirare used as the labels for the composite/parent tracks in the trackDb. However, these are required to be all lowercased (e.g. h3k27ac, bw, or clusters). This option allows one to specify the casing used for the short/long labels.
makeCbHub -d “Rad21 Olig2” bw/ track rad21 compositeTrack on shortLabel Rad21 longLabel Rad21 visibility dense autoScale group type bigWig ... track olig2 compositeTrack on shortLabel Olig2 longLabel Olig2 visibility dense autoScale group type bigWig ...
This command assumes that in bw/ (fileDir), there are two directories: rad21 and olig2, but it will use Rad21 and Olig2 as the shortLabel/longLabel for those composites in the trackDb.
Individual track labels: -s/--shortLabel and -l/–longLabel
The options -s/--shortLabel and -l/–longLabel allow you to control the short and long labels of the individual tracks in the composites. By default the script uses the file names as the labels, which, depending on how the files are named, can be pretty messy:
cd /hive/data/inside/cells/datasets/olg-eae-ms/eae-multiomics/hub makeCbHub bw/
Which results in:
...
track bw_P21208_1004_OPC_Ctr_RND1_peaks
parent bw on
shortLabel P21208_1004_OPC_Ctr_RND1_peaks
longLabel P21208_1004_OPC_Ctr_RND1_peaks
type bigWig 0.000000 160.000000
autoScale group
bigDataUrl bw/P21208_1004_OPC_Ctr_RND1_peaks.bw
visibility dense
...
However, if we rebuild the trackDb with these options:
cd /hive/data/inside/cells/datasets/olg-eae-ms/eae-multiomics/hub makeCbHub -s shortLabels.tsv -l acronyms.sorted.tsv bw/
Which outputs:
...
track bw_P21208_1004_OPC_Ctr_RND1_peaks
parent bw on
shortLabel OPC_Ctr
longLabel OPC_Ctr - Control oligodendrocyte precursor cells
type bigWig 0.000000 160.000000
autoScale group
bigDataUrl bw/P21208_1004_OPC_Ctr_RND1_peaks.bw
visibility dense
...
The shortLabels file contains two columns.
- file name
- short label
Here's the line from the shortLabels.tsv used in the example above:
P21208_1004_OPC_Ctr_RND1_peaks.bw OPC_Ctr
The longLabels file follows the same format as the 'acronymFile' setting that can be specified in the cellbrowser.conf (and will mostly likely be the same file). The two columns are:
- short label
- long label
Here's the line from the acronyms.sorted.tsv used in the example above:
OPC_Ctr Control oligodendrocyte precursor cells
Note: the shortLabel/longLabel files can be csv or tsv format and their file names need to end with csv or tsv (e.g. shortLabels.tsv).
Colors: -c/--colors
If a track doesn't include the 'color' setting, the Genome Browser colors that track black by default. The -c/–color option allows you to specify the color for each track. The file follows the same format as the 'colors' setting that can be specified in the cellbrowser.conf (and will mostly likely be the same file). The two columns are
- short label
- hexcode color (e.g. #834088), although RGB tuples (e.g. '131 64 136' or '131, 64, 136') are also accepted
Here are some commands so that you can see the setting in action:
cd /hive/data/inside/cells/datasets/olg-eae-ms/eae-multiomics/hub makeCbHub -s shortLabels.tsv -c ../colors.csv bw/
which outputs:
...
track bw_P21208_1004_OPC_Ctr_RND1_peaks
parent bw on
shortLabel OPC_Ctr
longLabel OPC_Ctr
type bigWig 0.000000 160.000000
autoScale group
color 131,64,136
bigDataUrl bw/P21208_1004_OPC_Ctr_RND1_peaks.bw
visibility dense
...
Here we needed to use the -s/--shortLabels setting in addition to the -c/--colors option because the short labels in the colors.csv match those in the shortLabels.tsv file. By default makeCbHub uses parts of the file name for the labels, meaning that if we omitted the -s/--shortLabels option the colors would show up as the labels makeCbHub is using wouldn't match those in the colors.csv file.
Here's an example line from colors.csv:
MOL12_EAE,#ECDD30
And the corresponding one from the shortLabels.tsv so that you can see the name overlap:
P21208_1005_MOL12_EAE_RND2_peaks.bw MOL12_EAE
Track description page: -f/--html
A track description page is there to briefly describe the data displayed by that track, any special display conventions, and contact info for the data creators.
Creating and adding a track description page is discussed in more detail in the BLAH section.
bigBed tracks: -b/--bbType
Most hubs you'll be building are going to be based on bigWig files. Occasionally, you may need to build one that includes a composite of bigBed files. The -b/--bbType option allows you to specify the bigBed type, e.g. 'bigBed 12 +', 'bigInteract', 'bigNarrowPeak'. The need for this option is because bigBed files are quite diverse. Many of the big* formats except for bigWig essentially use the bigBed format as a base but use a different 'type' in the trackDb to tell the Genome Browser how to interpret and display the different columns. It would be difficult to auto-detect the type from just the info in the file itself.
Here's an example using this option:
cd /hive/data/inside/cells/datasets/human-enhancer-atlas/hub makeCbHub --bbType bigNarrowPeak bb
Which outputs:
track bb
compositeTrack on
shortLabel bb
longLabel bb
visibility dense
autoScale group
type bigBed
track bb_Acinar
parent bb on
shortLabel Acinar
longLabel Acinar
type bigNarrowPeak
bigDataUrl bb/Acinar.bigBed
visibility dense
Using all these options together
Altogether, these options result in a solid track hub with user-friendly labels, colors, and a track description page. We'll skip the -b/--bbType since it's so rarely used. Here's a example you can run to see the results of combining these various options yourself:
cd /hive/data/inside/cells/datasets/olg-eae-ms/eae-multiomics/hub makeCbHub -s shortLabels.tsv -l acronyms.sorted.tsv -c ../colors.csv -f track_desc.html bw/
Which gets us:
track bw
compositeTrack on
shortLabel bw
longLabel bw
visibility dense
autoScale group
type bigWig
html track_desc.html
...
track bw_P21208_1004_OPC_Ctr_RND1_peaks
parent bw on
shortLabel OPC_Ctr
longLabel OPC_Ctr - Control oligodendrocyte precursor cells
type bigWig 0.000000 160.000000
autoScale group
color 131,64,136
bigDataUrl bw/P21208_1004_OPC_Ctr_RND1_peaks.bw
visibility dense
...
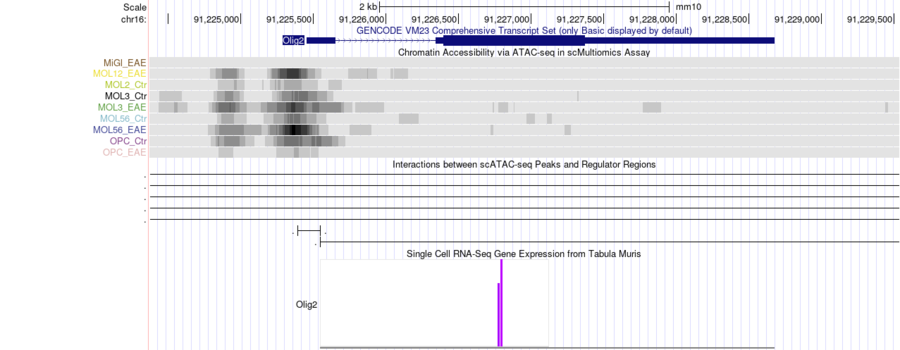
In this case, you may want to adjust the short/long labels for the parent track from bw to something like "Chromatin Accessibility", but small improvements like that are relatively easy. You can take a look at the hub.txt used this example was drawn from: https://cells.ucsc.edu/olg-eae-ms/eae-multiomics/hub/hub.txt, or take a look at the hub in the Genome Browser:
Building a hub
This section walks you through the process of creating a hub for a cell browser with tracks on a single assembly. If you are creating a hub with tracks on more than one assembly see the Hub Quick Start as it requires multiple configuration files.
File organization
makeCbHub assumes a certain directory structure when it creates the trackDb stanzas.
In your dataset directory, create a ‘hub’ directory where all of the hub-related files will live. In that hub directory, you will then create a directory for each composite/parent track (directory names should be all lowercase):
cb_dataset_dir/
|--> hub/
|--> track_set_A/
|--> track_A1.bw
|--> track_A2.bw
|--> etc…
|--> track_set_B
|--> track_B1.bw
|--> etc…
Dividing the individual tracks into composite/parent tracks will vary from dataset to dataset. For example, in the collection mouse-brain-cutandtag, individual tracks were divided into a composite track for each dataset in the collection (e.g. h3k27ac, h3k27me3, h3k27me3-cell-lines, h3k36me3, h3k4me3, olig2, rad21) as this was what was requested by the authors. In neuro-degen-atac, individual tracks were grouped according to their corresponding metadata field (e.g. broad-celltypes, clusters, neuronal-celltypes, neuronal-clusters). If you’re not sure how to group the tracks ask Max and/or the contributors.
Finally, it’s best to make symlinks to the track files in the orig directory to prevent the unnecessary duplication of large amounts of files. See human-enhancer-atlas/hub and fetal-chromatin-atlas/hub as examples, where the bigWigs alone were 212 GB and 138 GB, respectively. (/hive has a ton of storage, but it's good to not waste space unnecessarily.)
Track description page
You'll need to create a track description page for the hub. A track description page describes the data that people are looking at, any display conventions to keep in mind for that data (e.g. coloring), a contact email, and a reference to the paper(s). If you keep the page fairly high-level, you can use the same one for all of the tracks in the hub. Even if there are multiple composite/parent tracks in your hub, you can use the same description page for all of them since often the data are very similar.
You can use the Genome Browser template as a starting point. Additionally, it can be useful to look at the track description pages for other hubs in the Cell Browser:
olg-eae-ms/eae-multiomics: https://cells.ucsc.edu/olg-eae-ms/eae-multiomics/hub/track_desc.htmlolg-eae-ms/eae-atac: https://cells.ucsc.edu/olg-eae-ms/eae-atac/hub/track_desc.htmlneuro-degen-atacdataset: https://cells.ucsc.edu/neuro-degen-atac/track_desc.html (A good example of covering multiple composites with one page)
Once you have the page written, place it in the hub/ directory.
Coloring
For the "Display Conventions" section, a helpful tool that can quickly convert a colors file into an HTML list is generateColorHtml.
generateColorHtml colorFile
Where the colorFile is a two column file (csv or tsv). Column 1 is metadata value and Column 2 is a hex color.
For example, here is a peak inside the hea_colors.tsv for the human-enhancer-atlas dataset:
Adipocyte #002ea3 Airway Goblet Cell #25000d Alveolar Capillary Endothelial Cell #e1d0ff Alveolar Type 1 (AT1) Cell #0183f7 Alveolar Type 2 (AT2) Cell #00495a ... Vascular Smooth Muscle 1 #a60054 Vascular Smooth Muscle 2 #a37cff Ventricular Cardiomyocyte #00d2e8 Zona Fasciculata Cortical Cell #cad1ff Zona Glomerulosa Cortical Cell #ffe8ba
Paper Reference
A quick way to generate the HTML for a paper citation is by using getTrackReferences. You will need either the PubMed or the PubMedCentral IDs. You can run this script with multiple IDs at a time.
getTrackReferences ID_1 ID_2 ID_3
Hiding Contributor Email
It's good practice to encode the corresponding author emails using encodeEmail.pl. You can run this script on multiple emails at a time.
encodeEmail.pl email_1 email_2 email_3
Setting up your hub.txt
These lines will show up in any hub you create for the Cell Browser, though you have to adjust the values to be specific to your hub:
hub eae_multiomics shortLabel scATAC-seq of EAE Mice longLabel scATAC-seq of EAE Mice genomesFile genomes.txt email eneritz.agirre@ki.se descriptionUrl https://www.sciencedirect.com/science/article/pii/S0896627321010898 useOneFile on genome mm10
Put these lines in a file called hub.txt.
Breakdown of what to do for each setting in the first stanza:
hub- can be the same as the dataset short name (e.g. neuro-degen-atac)shortLabelandlongLabel- can be the same, though feel free to add more details to the longLabel if neededgenomesFile- will always be genomes.txtemail- should be the email of the contributordescriptionUrl- can point to the paper or to the track description page you createduseOneFile- will always be 'on'. The useOneFile option allows us to stick all of the hub configuration settings into one file.
After that, there should be a line break and then the genome setting. The value will be the name of the UCSC assembly the data was mapped to (e.g. hg38). You should confirm with the contributor the name of the genome assembly they used for their analysis. If they give you a non-UCSC name (e.g. GRCh38, GRCm39, etc.) you will need to map this to the UCSC assembly name. If it's a public dataset, you may be able to find this information in the methods section of the paper.
After that, you should add one more line break. This will ensure that the trackDb stanzas that you are going to add to the file in the next step will be properly separated from the genome line.
Running makeCbHub
Once you have your big* files in place, it's time to use makeCbHub to create the trackDb stanzas. In addition to the one required argument, you'll likely want to use these three as well:
-c/--colors-s/--shortLabels-f/--html
Beyond those three, you may also end up using the -d/--datasetList and -l/--longLabels options as well if needed.
Here's a set of example commands that you can use to create a set of trackDb stanzas using makeCbHub:
cd /hive/data/inside/cells/datasets/olg-eae-ms/eae-multiomics/hub makeCbHub -s shortLabels.tsv -l acronyms.sorted.tsv -c ../colors.csv -f track_desc.html bw
Which outputs:
track bw
compositeTrack on
shortLabel bw
longLabel bw
visibility dense
autoScale group
type bigWig
html track_desc.html
track bw_P21208_1004_OPC_Ctr_RND1_peaks
parent bw on
shortLabel OPC_Ctr
longLabel OPC_Ctr - Control oligodendrocyte precursor cells
type bigWig 0.000000 160.000000
autoScale group
color 131,64,136
bigDataUrl bw/P21208_1004_OPC_Ctr_RND1_peaks.bw
visibility dense
...
In reality, you'll be appending these lines to the end of your hub.txt file:
makeCbHub -s shortLabels.tsv -l acronyms.sorted.tsv -c ../colors.csv -f track_desc.html bw >> my_hub.txt
Files in /usr/local/apache/htdocs-cells
Once you have the big* files, hub.txt, and track description page all ready, then you will need to make symlinks to the files in /usr/local/apache/htdocs-cells.
First, navigate to your dataset's directory:
cd /usr/local/apache/htdocs-cells/${your_dataset_dir}
Then, make a 'hub' directory:
mkdir hub
Move into that directory, and create the proper symlinks:
cd hub
ln -s /hive/data./inside/cells/datasets/${your_dataset_dir}/hub/hub.txt
ln -s /hive/data./inside/cells/datasets/${your_dataset_dir}/hub/track_desc.html
ln -s /hive/data./inside/cells/datasets/${your_dataset_dir}/hub/track_A
etc ...
cellbrowser.conf settings
Now that you have everything in place in /usr/local/apache/htdocs-cells, you will need to add a few more settings to your cellbrowser.conf to have the links to the hub show up in the Cell Browser.
In your cellbrowser.conf, add these lines:
hubUrl="hub/hub.txt"
ucscDb="${ucsc_db_from_hub.txt}"
The value for ucscDb should match the value from the genome line in your hub.txt.
Rebuild your hub using cbBuild to get the links to the Genome Browser to appear in the Cell Browser, as can be seen for the cortex-dev dataset:
As you push data from cells-test to cells-beta and to cells, the hub files should be automatically copied from server to server.