Uploads by Hiram
From genomewiki
Jump to navigationJump to search
This special page shows all uploaded files.
| Date | Name | Thumbnail | Size | Description | Versions |
|---|---|---|---|---|---|
| 18:30, 28 April 2006 | PhenSort.gif (file) |  |
16 KB | Example phenotype | 1 |
| 17:37, 10 August 2006 | SmedleyCat.jpg (file) |  |
11 KB | Smedley the cat in the early days. Photo by Hiram Clawson, 1995. | 1 |
| 18:50, 10 August 2006 | StarTrekTOSTransporter.wav (file) | 223 KB | Testing wav file upload. Star Trek, The Original Series, Transporter sound. 10 seconds | 1 | |
| 19:00, 10 August 2006 | TestCustomTrack.data.txt.gz (file) | 2 KB | Testing file upload feature. This is a custom track test file demonstrating all track types currently (August 2006) supported by the browser custom tracks. | 1 | |
| 18:00, 11 June 2007 | CleanHgCentral pl.txt (file) | 2 KB | PERL cleaner script for userDb and sessionDb tables in hgcentral | 1 | |
| 16:33, 18 September 2007 | Mm8 mm9 exonsTo300.png (file) |  |
6 KB | Mouse mm8 and mm9 exon size histogram to size 300 nucleotides | 2 |
| 16:36, 18 September 2007 | Mm8 mm9.exonCount.png (file) |  |
5 KB | Mouse mm8 and mm9 exon count by size histogram, to size 30 nucleotides | 2 |
| 16:38, 18 September 2007 | Mm8 mm9.exonSize.png (file) |  |
5 KB | Mouse mm8 and mm9 exon size histogram, to size 50 nucleotides | 2 |
| 16:39, 18 September 2007 | Mm8 mm9.intronSize.png (file) |  |
5 KB | Mouse mm8 and mm9 intron size histogram, to size 50 nucleotides | 2 |
| 16:41, 18 September 2007 | Mm8 mm9.intronsTo170.png (file) |  |
5 KB | Mouse mm8 and mm9 intron size histogram, to size 170 nucleotides | 2 |
| 16:43, 18 September 2007 | Hg17 hg18.exonCount.png (file) |  |
5 KB | Human hg17 and hg18, exon count by size histogram, to size 30 nucleotides | 2 |
| 16:45, 18 September 2007 | Hg17 hg18.exonSize.png (file) |  |
5 KB | Human hg17 and hg18 exon size histogram, to size 50 nucleotides | 2 |
| 16:46, 18 September 2007 | Hg17 hg18.intronSize.png (file) |  |
5 KB | Human hg17 and hg18 intron size histogram to size 50 nucleotides | 2 |
| 16:46, 18 September 2007 | Hg17 hg18.intronsTo170.png (file) |  |
5 KB | Human hg17 and hg18 intron size to 170 nucleotides | 2 |
| 16:47, 18 September 2007 | Hg17 hg18 exonsTo300.png (file) |  |
6 KB | Human hg17 and hg18 exon size histogram, to a size of 300 nucleotides | 2 |
| 23:29, 18 September 2007 | Hg18KnownGenesSmallIntronsExonsCt.txt.gz (file) | 122 KB | Human Hg18 genome browser custom track to indicate unusually small introns and exons in the Known Genes version 3 procedure. These small exons and introns are used to maintain frame shifts in mRNA evidence that may be translation skipping or post translat | 1 | |
| 23:21, 10 October 2007 | Mm9 30way.gif (file) |  |
8 KB | Attempting to reinitialize this image to get it correct | 1 |
| 18:28, 30 October 2007 | PhastCons30way.png (file) |  |
5 KB | Mouse Mm9 30-way conservation phastCons histogram plot for the track data phastCons30way | 1 |
| 22:03, 3 December 2007 | Mm9Placental.png (file) |  |
5 KB | Mm9 alternative phastCons data histogram, Placentals, =mm9,rn4,cavPor2,oryCun1,hg18,panTro2,ponAbe2,rheMac2,calJac1,otoGar1,tupBel1,sorAra1,eriEur1,canFam2,felCat3,equCab1,bosTau3,dasNov1,loxAfr1,echTel1 | 3 |
| 22:04, 3 December 2007 | Mm9Euarchontoglires.png (file) |  |
5 KB | Mm9 alternative phastCons data histogram, Euarchontoglires, mm9,rn4,cavPor2,oryCun1,hg18,panTro2,ponAbe2,rheMac2,calJac1,otoGar1,tupBel1 | 3 |
| 21:40, 7 December 2007 | UserCount.png (file) | 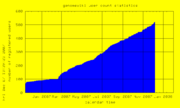 |
9 KB | Genomewiki User Count graph | 1 |
| 21:53, 7 December 2007 | PageCount.png (file) | 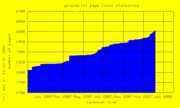 |
8 KB | Genomewiki page count graph | 1 |
| 21:41, 17 March 2008 | TestOOPresent.ppt (file) | 11 KB | Testing ppt upload | 1 | |
| 22:47, 13 May 2008 | EmbedURL php.txt (file) | 3 KB | UCSC modified EmbedURL extension code php file. Adds scrolling and frameborder options, and defaults width and height to 100% and 500 | 1 | |
| 05:15, 12 July 2008 | CSHL 2003 Jim Kent.ppt (file) | 1.52 MB | CSHL June 2003 "Evolution and the Santa Cruz Genome Browser" presentation by Jim Kent. Recent new functions in the genome browser, using the chain and net process to highlight evolutionary processes, a new rat genome assembly with 3-way human-mouse-rat mu | 1 | |
| 06:14, 13 July 2008 | SoftwareSermon2002 Jim Kent.ppt (file) | 113 KB | Genecats October 2002 - "Software Sermon" - software development philosophy and guidelines, Jim Kent. | 1 | |
| 06:43, 13 July 2008 | CompBioOct2000 Jim Kent.ppt (file) | 45 KB | Description of the genome browser software architecture, the early days. Jim Kent | 1 | |
| 14:39, 23 July 2008 | ISMB2008 UCSC.ppt (file) | 5.35 MB | ISMB 2008 Toronto - Intelligent Systems for Molecular Biology - presentation by Hiram Clawson. An example table browser intersection is performed with all the steps indicated. | 1 | |
| 22:00, 6 August 2008 | ISMB2008 genecats summary.ppt (file) | 3.8 MB | Hiram's summary of ISMB 2008 Toronto Ontario, July 2008 | 1 | |
| 17:32, 16 December 2008 | Baertsch-code-talk.ppt (file) | 50 KB | BME230 lecture by Robert Baertsch, Winter 2008, 10 January 2008, genome browser introduction and kent source tree preview | 1 | |
| 17:47, 16 December 2008 | Metcalf.Richardson.2008-10-15.pdf (file) | 3.26 MB | Jake Metcalf and Sarah Richardson, Genecats presentation 15 Oct 2008, ethics discussion of Bruce T. Lahn controversy from U. Chicago. | 1 | |
| 18:39, 16 December 2008 | ISMB2007TripReportGenecats.ppt (file) | 4.42 MB | Rachel Harte genecats presentation summary from ISMB 2007, Vienna Austria | 1 | |
| 18:57, 16 December 2008 | Kent allJoiner.ppt (file) | 41 KB | Jim Kent, genecats 05 July 2008 - the all.joiner file which describes the database schema relationships for the UCSC genome browser | 1 | |
| 19:02, 16 December 2008 | Kent parasolArchitecture.ppt (file) | 60 KB | Jim Kent genecats presentation June 2008, Parasol super computer job management system review | 1 | |
| 18:44, 17 December 2008 | Kent decodingEncode.ppt (file) | 4.24 MB | Jim Kent at the Computational Systems Bioinformatics Conference, Stanford CA, 27 Aug 2008. Encode, the genome browser, the table browser, the source code, a variety of subjects are discussed. | 1 | |
| 18:55, 17 December 2008 | MouseAnnotAndCCDSVisitReport041608.ppt (file) | 2.34 MB | Rachel Harte summarizing Mouse Genome Annotation meeting and RefSeq/CCDS groups, NIH March 2008. | 1 | |
| 22:05, 17 December 2008 | Flicek 11 DEC 2008.pdf (file) | 3.41 MB | Paul Flicek, EBI, presenting 1,000 genomes project status | 1 | |
| 17:42, 19 December 2008 | FrankJacobs181208.ppt (file) | 29.16 MB | Frank Jacobs from Rudolf Magnus Institute of Neuroscience, University of Utrecht, The Netherlands, "Pitx3 outSMRTs Nurr1 in dopamine neuron terminal differentiation" | 1 | |
| 17:51, 8 January 2009 | BME230 Winter 2009.ppt (file) | 5.88 MB | Hiram Clawson, BME 230 lecture, 08 January 2009, introduction to the genome browser and programming with the kent source tree. | 2 | |
| 18:27, 25 March 2009 | Teal Day 05.jpg (file) |  |
1.02 MB | Trying to recover a mysterious bogus file directory link | 1 |
| 18:46, 29 July 2009 | SvnForGb.pdf (file) | 171 KB | Mark's presentation to 2009-07-29 genecats, review of SVN subversion and how it could replace CVS | 1 | |
| 22:28, 17 September 2009 | MultAlign.jpg (file) |  |
14 KB | Example tool tip pop up image | 1 |
| 20:46, 14 October 2009 | Genecats 2009 10 14.ppt (file) | 141 KB | Building the kent source tree and a UCSC genome browser assembly, Genecats 2009-10-14 Hiram Clawson | 1 | |
| 20:46, 14 October 2009 | Genecats 2009 10 14.pptx (file) | 86 KB | Building the kent source tree and a UCSC genome browser assembly, Genecats 2009-10-14 Hiram Clawson | 1 | |
| 22:14, 14 October 2009 | MonsatoIntroduction Oct2009.pdf (file) | 8.37 MB | Montsanto Introduction, Genecats 14 Oct 2009 | 1 | |
| 19:35, 30 November 2009 | PhyloP46way.histogram.png (file) |  |
6 KB | 46-way conservation track on human genome browser hg19. phyloP histogram data for all 46 vertebrates | 1 |
| 19:36, 30 November 2009 | PhyloP46wayPlacental.histogram.png (file) |  |
6 KB | 46-way conservation track on human genome browser hg19. phyloP histogram data for all placental mammal subset | 1 |
| 19:37, 30 November 2009 | PhyloP46wayPrimates.histogram.png (file) |  |
6 KB | 46-way conservation track on human genome browser hg19. phyloP histogram data for all primates subset | 1 |
| 19:24, 1 December 2009 | PhastCons46way.histogram.png (file) |  |
5 KB | 46-way conservation track on human genome browser hg19. phastCons histogram data for all 46 vertebrates | 1 |
| 19:24, 1 December 2009 | PhastCons46wayPlacental.histogram.png (file) |  |
5 KB | 46-way conservation track on human genome browser hg19. phastCons histogram data for all placental mammal subset | 1 |