Encode scenarios: Difference between revisions
(needed catergory to be in CAPs) |
(added two more scenarios and put into table for formatting.) |
||
| Line 1: | Line 1: | ||
{| class="wikitable" border="0"|- | |||
|- | |||
| | |||
== Tier 1 Cell Lines == | == Tier 1 Cell Lines == | ||
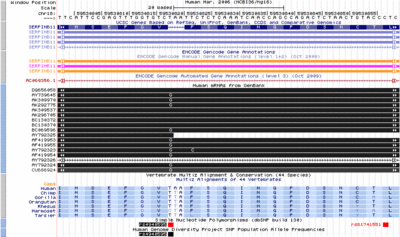
[[Image:EncodeKent1.gif|400px|thumb|left|Tier 1 cell lines]] | |||
[[Image:EncodeKent1. | |||
[http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=Kuhn&hgS_otherUserSessionName=hg18_encodeKent1 Launch Browser] | [http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=Kuhn&hgS_otherUserSessionName=hg18_encodeKent1 Launch Browser] | ||
| Line 9: | Line 10: | ||
which shows the level of expression of the gene EWSR1, the | which shows the level of expression of the gene EWSR1, the | ||
histone mark, H3K4Me, which is associated with promoters; DNAseI | histone mark, H3K4Me, which is associated with promoters; DNAseI | ||
hypersensitivity, which is associated with regulatory regions in general; and ChIP-seq showing levels of occupancy by the transcription factor c-Fos. | hypersensitivity, which is associated with regulatory regions in general; | ||
and ChIP-seq showing levels of occupancy by the transcription factor c-Fos. | |||
contributed by Jim Kent. | |||
|- | |||
| | |||
== Premature Stop in Reference Assembly == | |||
[[Image:EncodeKent2.gif|400px|thumb|left|Premature stop in reference assembly]] | |||
[http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=Kuhn&hgS_otherUserSessionName=hg18_encodeKent2 Launch Browser] | |||
A premature stop codon that is found in the reference genome and about half of people of European descent. The UCSC Genes track is forced to skip the codon. | |||
contributed by Jim Kent. | |||
|- | |||
| | |||
== Multiple Histone Marks == | |||
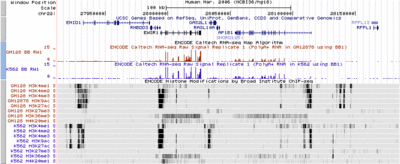
[[Image:EncodeKent3.gif|400px|thumb|left|Multiple histone marks.]] | |||
[http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=Kuhn&hgS_otherUserSessionName=hg18_encodeKent3 Launch Browser] | |||
All available histone ChIP-seq data on the two tier one cell lines. | |||
H3K4me1 is associated with enhancers and to an extent with promoters, | |||
H3K4me3 with promoters, and H3K36me3 with transcribed regions. | |||
contributed by Jim Kent. | |||
|- | |||
| | |||
== Template == | == Template == | ||
| Line 19: | Line 55: | ||
Description of scenario. | Description of scenario. | ||
contributed by . | |||
|} | |||
Revision as of 02:19, 13 March 2010
Tier 1 Cell LinesBasic ENCODE data on the Tier 1 cell lines GM128, a lymphoid cell line and K562, a myeloid line. The tracks include RNA-seq which shows the level of expression of the gene EWSR1, the histone mark, H3K4Me, which is associated with promoters; DNAseI hypersensitivity, which is associated with regulatory regions in general; and ChIP-seq showing levels of occupancy by the transcription factor c-Fos. contributed by Jim Kent.
|
Premature Stop in Reference AssemblyA premature stop codon that is found in the reference genome and about half of people of European descent. The UCSC Genes track is forced to skip the codon.
|
Multiple Histone MarksAll available histone ChIP-seq data on the two tier one cell lines. H3K4me1 is associated with enhancers and to an extent with promoters, H3K4me3 with promoters, and H3K36me3 with transcribed regions.
|
TemplateDescription of scenario.
|