Encode scenarios: Difference between revisions
(added to encode category) |
mNo edit summary |
||
| (2 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{| class="wikitable" border="0"|- | |||
|- | |||
| | |||
== Tier 1 Cell Lines == | == Tier 1 Cell Lines == | ||
[[Image:EncodeKent1.gif|400px|thumb|left|Tier 1 cell lines]] | |||
[[Image:EncodeKent1. | [http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=Kuhn&hgS_otherUserSessionName=hg18_encodeKent1 Launch active Browser at this location] | ||
[http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=Kuhn&hgS_otherUserSessionName=hg18_encodeKent1 Launch Browser] | |||
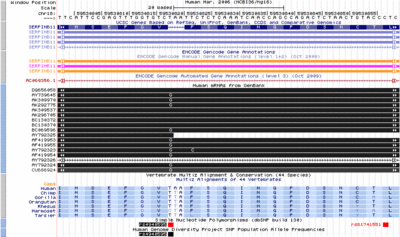
Basic ENCODE data on the Tier 1 cell lines GM128, a lymphoid | Basic ENCODE data on the Tier 1 cell lines GM128, a lymphoid | ||
| Line 9: | Line 10: | ||
which shows the level of expression of the gene EWSR1, the | which shows the level of expression of the gene EWSR1, the | ||
histone mark, H3K4Me, which is associated with promoters; DNAseI | histone mark, H3K4Me, which is associated with promoters; DNAseI | ||
hypersensitivity, which is associated with regulatory regions in general; and ChIP-seq showing levels of occupancy by the transcription factor c-Fos. | hypersensitivity, which is associated with regulatory regions in general; | ||
and ChIP-seq showing levels of occupancy by the transcription factor c-Fos. | |||
contributed by Jim Kent. | |||
|- | |||
| | |||
== Premature Stop in Reference Assembly == | |||
[[Image:EncodeKent2.gif|400px|thumb|left|Premature stop in reference assembly]] | |||
[http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=Kuhn&hgS_otherUserSessionName=hg18_encodeKent2 Launch active Browser at this location] | |||
A premature stop codon that is found in the reference genome and about half of people of European descent. The UCSC Genes track is forced to skip the codon. | |||
contributed by Jim Kent. | |||
|- | |||
| | |||
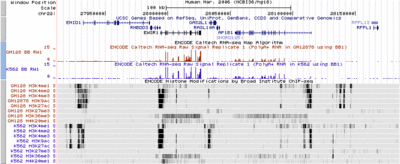
== Multiple Histone Marks == | |||
[[Image:EncodeKent3.gif|400px|thumb|left|Multiple histone marks.]] | |||
[http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=Kuhn&hgS_otherUserSessionName=hg18_encodeKent3 Launch active Browser at this location] | |||
All available histone ChIP-seq data on the two tier one cell lines. | |||
H3K4me1 is associated with enhancers and to an extent with promoters, | |||
H3K4me3 with promoters, and H3K36me3 with transcribed regions. | |||
contributed by Jim Kent. | |||
|- | |||
| | |||
== Template == | == Template == | ||
[[Image:image.jpg|400px|thumb|left|text below image]] | [[Image:image.jpg|400px|thumb|left|text below image]] | ||
[http://urlToSession Launch Browser] | [http://urlToSession Launch active Browser at this location] | ||
Description of scenario. | Description of scenario. | ||
contributed by . | |||
|} | |||
Latest revision as of 17:40, 12 April 2010
Tier 1 Cell LinesLaunch active Browser at this location Basic ENCODE data on the Tier 1 cell lines GM128, a lymphoid cell line and K562, a myeloid line. The tracks include RNA-seq which shows the level of expression of the gene EWSR1, the histone mark, H3K4Me, which is associated with promoters; DNAseI hypersensitivity, which is associated with regulatory regions in general; and ChIP-seq showing levels of occupancy by the transcription factor c-Fos. contributed by Jim Kent.
|
Premature Stop in Reference AssemblyLaunch active Browser at this location A premature stop codon that is found in the reference genome and about half of people of European descent. The UCSC Genes track is forced to skip the codon.
|
Multiple Histone MarksLaunch active Browser at this location All available histone ChIP-seq data on the two tier one cell lines. H3K4me1 is associated with enhancers and to an extent with promoters, H3K4me3 with promoters, and H3K36me3 with transcribed regions.
|
TemplateLaunch active Browser at this location Description of scenario.
|