Opsin evolution: key critters: Difference between revisions
Tomemerald (talk | contribs) No edit summary |
Tomemerald (talk | contribs) No edit summary |
||
| Line 48: | Line 48: | ||
No hagfish opsins have been sequenced; no genome project is scheduled. Even if hagfish imaging opsins are mostly gone, other ciliary and rhabdomeric opsins could be quite informative. Hagfish may have information about a critical era in deuterostome imaging opsin evolution. | No hagfish opsins have been sequenced; no genome project is scheduled. Even if hagfish imaging opsins are mostly gone, other ciliary and rhabdomeric opsins could be quite informative. Hagfish may have information about a critical era in deuterostome imaging opsin evolution. | ||
* <span style="color: #990099;">Hypsibius (water bear) tardigrade no opsins yet</span> | |||
A 5x genome project for Hypsibius dujardini, a phylum of microscopic ecdysozoan was approved in July 2007 but Broad has not yet begun trace reads on the small 70 mbp genome (suggesting densely spaced genes with small introns as this is not likely highly derived). It could prove very useful for opsins as [http://www.genome.gov/Pages/Research/Sequencing/SeqProposals/EcdysozoaProposalFinalPDF.pdf tardigrades] are basal to all of Arthropoda and so shed light on that last common ancester. In fact with accompanying centipede, horseshoe crab, amphipod, and priapulid genomes, the whole ecdysozoan ancester will be accessible. | |||
The only known fossil specimens are found in Siberian mid-Cambrian deposits and much later amber. The older fossils have three pairs of legs rather than four, a simplified head morphology, and no posterior head appendages and probably represent a stem group of extant tardigrades. Aysheaia from the Burgess Shale might be related to tardigrades. | |||
Nothing is currently known about photoreception or opsins in tardigrades -- or even if they have eyes. However it looks like we can expect some rhabdomeric opsins at the minumum in front of these pigment cups. However the current GSS and EST collections (about 6000 sequences) do not currently contain any convincing matches using various rhabdomeric and ciliary opsins as tblastn queries. Tardigrade photos and movies provided by [http://www.bio.unc.edu/faculty/goldstein/lab/willow.html Goldstein Lab] | |||
[[Image:tardi.png|center|]] | |||
* <span style="color: #990099;">Ixodes (tick)</span> | * <span style="color: #990099;">Ixodes (tick)</span> | ||
| Line 91: | Line 102: | ||
* <span style="color: #990099;">Stronglyocentrotus (sea urchin) echinoderm opsins</span> | * <span style="color: #990099;">Stronglyocentrotus (sea urchin) echinoderm opsins</span> | ||
* <span style="color: #990099;">Saccoglossus (acorn worm) | * <span style="color: #990099;">Saccoglossus (acorn worm) hemichordate opsins</span> | ||
* <span style="color: #990099;">Xenoturbella plus Convoluta no opsins yet</span> | * <span style="color: #990099;">Xenoturbella plus Convoluta no opsins yet</span> | ||
* <span style="color: #990099;">Schmidtea (planaria) opsins</span> | * <span style="color: #990099;">Schmidtea (planaria) opsins</span> | ||
* <span style="color: #990099;">Platynereis (polychaete) lophotrochozoan opsins</span> | * <span style="color: #990099;">Platynereis (polychaete) lophotrochozoan opsins</span> | ||
Revision as of 20:09, 25 November 2007
Some species such as drosophila have lost all ciliary opsins [ref] -- clearly they are not essential for a successful visually complex flying insect with 5-color vision and circadian rhythm. Bees, annelids, and mammals retain ciliary opsins so we know this must be the ancestral bilateran state state. This predicts ciliary opsins in cnidaria and indeed one was just found in cnidaria. One sees the importance of complete genomes here (versus transcripts or immunostained sections): absence of ciliary opsin evidence in a genome is truly evidence of ciliary opsin absence.
When the eye is reduced to a single pigment cell backing a single photoreceptor cell, the opsin of that species will be expressed only in one cell of the entire body. In this situation, the opsin may never show up in transcript collections, even with subtraction of common ones.
Vertebrates could never have evolved cilliary opsin vision had the bilateran ancestor possessed the limited opsin repertoire of fruit fly. Thus most pressing question is -- assuming rhabdomeric opsins were thoroughly entrenched in the earliest imaging eyes and photoreception systems -- what kept ciliary opsins around in early bilatera (and even cnidaria) so that they could later be co-opted for ciliary opsin-based vision? We could also ask why vertebrates did not stay on the rhabdomeric track of early deuterostomes but instead underwent this profound switch to the 'untested' ciliary track. It is not at all clear what advantages ciliary offers over rhabdomeric -- ever miss swatting a fly?
Fossil dna does not go back nearly this far back, the nearly non-existent fossil record of soft body parts is unhelpful, and transcripts plus genomes of living species are 450-550 million years removed from the crucial ancestral chain of events. For example, transcript-labelled thin sections of photoreceptor systems in modern amphioxus only speak to the current situation, as does extraction of opsin genes from the new assembly, and speak to the history only by inference. The situation today may be seriously different from the ancestor in terms of both innovations and losses. And that history is not necessarily the most parsimonius (even though we will often assume that).
It's worth expanding on this perpetual source of confusion by emphasizing here too that contemporary tunicates, lancelets, and lamprey are not ancient, ancestral, antiquated, archaic, character-retaining, dead-end, failed experiments, frozen in time, genetically stationary, living fossil, primitive, primordial, relic, or retro species. They're full modern -- the tree of life is right-justified. Indeed their genes, regulatory signalling systems, and enzymes may be more finely honed than mammals because of more rapid evolution attributable to larger effective population sizes, reproductive mode, generation time, and marine selective predatory pressures.
However we can hope that ancestral character traits will still be reflected to some extent in these earlier diverging species and that with enough complete opsin repertoires from taxonomically appropriate species, the ancestral genes and even visual systems can be reconstructed at key nodes on the phylogenetic tree. The story describing the evolution of the human eye then amounts to describing the status at these successive nodes and perhaps interpolating between them. There are definitely limits to knowledge here as extant metazoans provide only 35 nodes between sponge and human -- gaps between nodes may average 30 million years but can seriously exceed that. This is offset by the occasional proposal of new deuterostome branches (Xenoturbella, Convoluta).
The ideal set of genomes needed to study the evolution of the metazoan eye is only partly completed, underway, or even proposed. In some cases, the genome size of clade representatives is so large (eg lungfish at 25x human) the species may never be sequenced, though opsin transcripts could still be obtained. In others, the rate of evolution has been so fast so long that very little information about photoreception at ancestral nodes can remain (eg Oikopleura). Hagfish opsins, which would conveniently break up the crucial lamprey long branch, are not available at GenBank but here the animal has adopted a deep water (dark) habitat, meaning that its cone opsin genomic repertoire will be highly reduced, if not gone entirely, in its markedly degenerated eyes. (Its other opsins could still be informative.)
Model organism choices do not always coincide with genome sequenceability, transcriptome projects, nor (worst of all) with slow-evolving less derived species. Finally, most sequencing speaks to narrow anthropocentric interests, whereas the more broadly conceived sequencing need is greatest farther back (to break up branches). The evolution of the eye needs a rather different portfolio of genomes than a typical disease gene because of the earlier intrinsic timing of the innovative events. In fact, one product of the investigation here is to spell out these needed genomes. Of course one obvious genome choice is the cubomedusan jellyfish Carybdea marsupialis with its 24 eyes of 6 types.
It's worth reviewing genome status and recent experimental literature on key species. While abstracts are readily available at PubMed, access to free full text is unpredictable, so those links are collected when available. It suffice to reference only recent articles because they cite the earlier literature and citation in turn of their paper are collected by Google Scholar (or AbstractsPlus at PubMed). Most opsin sequences in the Opsin evolution reference sequence collection have a PubMed accession as a field their fasta header database; those can simply be compiled to an active link that opens all of them in one PubMed window.
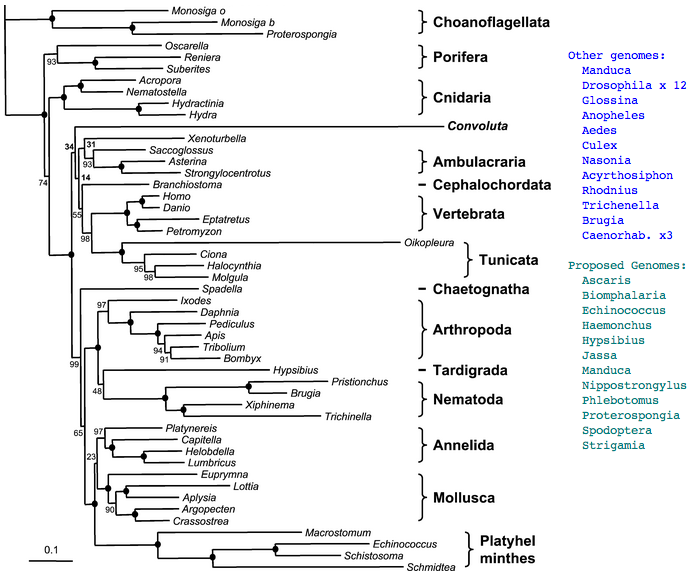
Figure adapted from: Acoel Flatworms Are Not Platyhelminthes: Evidence from Phylogenomics Hervé Philippe et al PLoS ONE. 2007 Aug 8
- Callorhinchus (elephantshark) chondrichthyes opsins
Five ray-finned fish genomes are available but these have major lineage-specific expansions and are quite derived. Some sequences are available from lobed-finned fish and coelocanth genome has been proposed. This makes the preliminary genome assembly of the much earlier diverging Callorhinchus (oft-misspelled) and skate transcipts very special because it is the "last stop" before lamprey.
This large-eyed cartilaginous fish lives to depths to 200m on the continental shelf of southern Australia and New Zealand but migrates into coastal estuaries to lay egg cases (lower image) in sand and muddy substrates. The distinctively-shaped egg cases are sometimes found washed ashore after storms. They are up to 25cm long, 10cm wide, and take up to eight months to hatch. The one studied member of the genus has a vitamin A1-based photopigment with maximum absorbance at 499 nm presumably adapted to its overall photic environment.
I made an exhaustive search of the WGS and Trace divisions of GenBank on 5 Nov 2007, recovering many complete exons but most fragmentary genes. The opsin classifier can easily place these fragments. Overall, Callorhinchus appears to have a full complement of vertebrate opsin genes. The exceptions are RHO2, SWS1, SWS2 (oddly also missing in skate and dogfish ESTs) apparently leaving elephantshark with only RHO1 and LWS rod/cone pigments. Parietopsin was also missing. Two encephalopsin- and two melanopsin-class opsins were found. The RGR, peropsin, and neuropsin genes will prove important in better determining their overall gene tree placement (which an October 2007 opsin phylogeny paper placed deeply within rhabdopsins).
- Petromyzon (lamprey) agnathan opsins
Lamprey are the favored outgroup to jawed vertebrates. Geotria and Petromyzon split 280-220 myr ago (helpful in breaking a long branch in half) whereas Lethenteron-Petromyzon was much later 20 myr (unhelpful, too close). Their photoreceptor systems have been studied in considerable depth.
Geotria australis has a full complement of imaging opsins LWS, SWS1, SWS2, RHO2, and RHO1 as well as the other major opsin classes. This implies that the ancestral vertebrate possessed photopic (bright light) cone-based vision with the potential for pentachromacy, circadian rhythm, pupilary reflex, and pineal and parapineal functions. Photoreceptor morphology and spectral sensitivity can change during various phases of the lamprey lifecycle, a lineage-specific complication that does not concern us.
The lamprey genome project has stalled, despite accruing an impressive 19 million traces. It seems a 16 kbp retroposon has expanded enormously, making assembly all but impossible. However new sequencing technologies make an immense collection of cDNA affordable. This would permit a pseudo-assembly of exons flanked by some at least genomic dna. That is, transcripts are aligned into the trace archives to obtain non-coding context for exons. This allows some topics to be studied (intron retention, invariant non-coding) but not others (upstream regulatory, chromosomal gene order). Labelling techniques such as FISH could provide some linkages but not gene-level order; perhaps this could be done at the level of BACs using exons from all possible gene pairs. Alternatively, the genome of Geotria might present fewer issues.
The high lamprey trace coverage is very helpful to this project. Because most of the opsin sequences are from non-genomic species, the intron structures were not known. I've mapped all GenBank opsin data for Geotria and Lethenteron into Petromyzon using these traces, not only obtaining a nearly full spectrum of new sequences but also sequences parsed for intron breaks and phase. Further, opsin classes not available in other lamprey were collected using chondrichthyes genes as query. In all cases, intron placement remains perfectly conserved from lamprey to mammal, indeed to amphioxus with some complications. These could help validate orthology candidates in earlier species, especially to clarify the presumably ancient but still cryptic origin of imaging opsins. Intron gain and loss is rare in many lineages -- the vast majority of human introns are conserved back to Urbilatera, indeed to sponge.
- Eptatretus (hagfish) agnathan opsins
Hagfish, after decades of back-and-forth, are now sistered with lamprey, news not accepted yet by the Taxonomy division of GenBank. However including rogue taxa such as Oikapleura can severely skew results in molecular phylogeny studies. Monophyly of Cyclostoma is unfortunate if true -- an earlier node would very much help in understanding the origin of the eye. It would be better to put aside tree topology until rare genomic events can be developed and simply procede with opsin sequencing.
Jawless fish first appeared in the Ordovician. Hagfish and lamprey split well after the Cambrian, roughly 430 myr ago according to molecular clocks. That's a time span comparable to divergence of human from shark. The oldest fossil hagfishes are Late Carboniferous (330 myr). The two extant hagfish groups split some 75 myr ago (human from mouse). Only recently has it been possible to obtain hagfish eggs and embryos and revisit the neural crest issue. Hagfish experienced an extra round of HOX gene expansion, undercutting both HOX cluster copy number as a hallmark of supposed vertebrate body plan innovation and premature speculation on 1R and 2R whole genome duplication.
Hagfish are nocturnal in aquaria and deep-sea in their natural habitat -- a new Eptatretus species was even captured at a hydrothermal vent. This lifestyle is not conducive to a well-preserved imaging opsin portfolio, though hagfish still have circadian rhythm (based in the preoptic nucleus) and dermal photoreceptors though no pineal gland. The non-imaging paired eyes lack cornea, lens, vitreous body, and extrinsic eye muscle but nonetheless the retina and optic nerve react with opsin antibody. The eyes are larger in Eptatretus than in Myxine, where they are partly covered by the trunk musculature. However 1.3 mm is still quite small for an eye. After comparison of all extant genera, Fernholm and Holmberg concluded in 1975 that the hagfish eyes are secondarily degenerated from more conventional eyes adapted for shallow water (for example an early lens placode disappears). The comparative anatomy of hagfish eyes has an excellent discussion in The Biology of Hagfishes (JM Jorgensen), pages 542-543 available by google book search.
No hagfish opsins have been sequenced; no genome project is scheduled. Even if hagfish imaging opsins are mostly gone, other ciliary and rhabdomeric opsins could be quite informative. Hagfish may have information about a critical era in deuterostome imaging opsin evolution.
- Hypsibius (water bear) tardigrade no opsins yet
A 5x genome project for Hypsibius dujardini, a phylum of microscopic ecdysozoan was approved in July 2007 but Broad has not yet begun trace reads on the small 70 mbp genome (suggesting densely spaced genes with small introns as this is not likely highly derived). It could prove very useful for opsins as tardigrades are basal to all of Arthropoda and so shed light on that last common ancester. In fact with accompanying centipede, horseshoe crab, amphipod, and priapulid genomes, the whole ecdysozoan ancester will be accessible.
The only known fossil specimens are found in Siberian mid-Cambrian deposits and much later amber. The older fossils have three pairs of legs rather than four, a simplified head morphology, and no posterior head appendages and probably represent a stem group of extant tardigrades. Aysheaia from the Burgess Shale might be related to tardigrades.
Nothing is currently known about photoreception or opsins in tardigrades -- or even if they have eyes. However it looks like we can expect some rhabdomeric opsins at the minumum in front of these pigment cups. However the current GSS and EST collections (about 6000 sequences) do not currently contain any convincing matches using various rhabdomeric and ciliary opsins as tblastn queries. Tardigrade photos and movies provided by Goldstein Lab
- Ixodes (tick)
The genome project was completed long ago but has experienced a multi-year bottleneck in assembly release and publication. However contigs built from 19.4 million traces should become available to tblastn of the GenBank "wgs" division by late 2007. Ixodes has a very conservative genome (regretably 2.1 gbp in size), seemingly far less derived than drosophilids in matters such as intron, gene retention, and protein sequence conservation. This, in conjunction with the helpful phylogenetic position of chelicerate outgroup to the many insect genomes, has improved prospects for reconstructing the ancestal opsin repertoire of Arthropoda and eventually Protostomia and UrBilatera.
A large collection of annotated Ixodes ESTs is available at the DFCI Gene Index of which 3 are marked up (2 wrongly) as opsins. Using the Opsin Classifier, I recovered the full length gene for the first of these TC19272 on 24 Nov 07, intronated the transcript at the Trace Archives (4 introns, superb coverage), and added it to the classifier fasta collection as RHAB1_ixoSca. It classifies with rhabdomeric opsins (ie with deuterostome melanopsins) with a very respectable 57% maximal percent protein identity. The second and third intron have classical ancestral position (following GWSR and LAK) and phase (2 and 0). Synteny awaits assembly of large contigs -- adjacent exons are not spanned by single traces. An apparent ciliary opsin fragment in Ixodes was located using that of Platynereis dumerilii as probe, it is stored as CILI_ixoSca but needs further analysis.
- Daphnia (water flea)
An 8.7x genome assembly was released in July 2007 at JGI with further support at wFleaBase. This crustacean provides a potentially important outgroup to insects (together forming Pancrustacea). However the opsin story, summarized in a meeting abstract is an embarrassment of riches, not conducive to deducing ancestral arthropod genome content. The total number of opsin genes came in at 37, comprised of 22 rhabdomeric opsins (mostly long wavelength), 7 ciliary opsins (pteropsins), and 8 in a novel family without close affiliates. This seems excessive but Daphnia has ommatidia (compound eyes), circadian rhythms, and a need to assess water turbidity and depth. Planned in situ hybridization studies may illuminate biological roles of these opsins. The pteropsins are probably of most interest here.
Gene models have not been submitted yet to GenBank but are likely extractable by text query at wFleaBase. What is needed here however is not the clutter of 37 sequences but their collapse into UV, blue, long, pteropsin, and novel ancestral representatives. This would remove 'noise' from lineage-specific expansions. The intron structure could provide very important support to classification schemes.
The expansions may have arisen through retroprocessing (rather than segmental duplication) of a few master exonic genes, which would then be the orthologs to other arthropod opsins. Indeed the intronation pattern -- typically far more deeply conserved than protein sequence -- could link pteropsins more convincingly to lophotrochoan and deuterostome opsins than alignments with percent identities in the 20's.
This blast twilight zone is especially dangerous for photoreceptor opsins because they are embedded in much larger gene family of generic rhodopsin and GPCR which share many structural and signaling properties. A slowly evolving generic rhodopsin might well score higher than fast evolving photoreceptor opsins. Gene expansions are noted for markedly enhanced rates as copies neo- or subfunctionalize. The generic rhodopsin might also share diagnostic residues through convergence at least at the level of statistical signficance ambiguity. Consequently intron location/phase and synteny can provide important backup.
The synteny circle surviving at this phylogenetic depth will be local (optimistically Pancrustacean). That is, the blue opsin of Daphnia might in synteny with Drosophila (ie establish orthology) but not to Platynereis ciliary opsin much less any vertebrate opsin (eg encephalopsin). This could be remedied to some extent by ancestral gene order reconstruction. The degree to which synteny can contribute to validating orthology relations within opsins is not currently known.
- Carybdea marsupialis (jellyfish) cnidarian no opsins yet
Cnidarians are the earliest diverging invertebrates with multicellular light-detecting organs, called ocelli. Photodetectors include simple eyespots, pigment cups, complex pigment cups with lenses, and camera-type eyes with a cornea, lens, and retina. These remarkable eyes are located on sensory clubs called rhopalia with four lining the bell of Each houses six eyes: a pair of pit ocelli, a pair of slit ocelli, and two unpaired lens eyes with counterparts to cornea, cellular lens and retina of ciliated photoreceptors. Anatomically, ocelli have bipolar sensory photoreceptor cells interspersed among nonsensory pigment cells with the apical end making the light-receptor and the basal end forming an axon that synapses with second-order neurons to form what amounts to ocular nerves.
The spectral sensitivity of neritic (near-shore) lens eyes of a box jellyfish, Tripedalia cystophora recently considered by MM Coates et al was interpreted as a single vitamin A-1 based opsin with peak sensitivity near 500 nm (blue-green). However nothing was sequenced.
The most striking jellyfish from the perspective of a complex set of eyes is Carybdea marsupialis, as reviewed by VJ Martin. Antibody studies based on vertebrate cone/rod opsins are doubtful because of cross reactivity to generic GPCR proteins; again no opsins have been sequenced yet.. This would make a great genome to study provided the retroposon and base composition are not unwieldy. Nematostella and Hydra, whatever their other genomic merits, sits in the Anthozoa and Hydrozoa respectively, types of cnidarian lacking elaborate visual systems.
Vision has roles in the reproduction and feeding of cubomedusae which can find each other and chase, catch, and eat teleost fish. A patch of Pelagia nocticula 10 square miles in extent and 35 feet deep recently destroyed a salmon farm off Northern Ireland.
- Amphimedon (sponge) and earlier metazoan lack opsins
Sponges lie at the base of multicellular animals. They are not noted for eyes. However demosponge larva do exhibit phototaxis (shadow seeking under coral rubble) but the action spectrum is reportedly a better fit to a flavin or carotenoid chromophore. The genome of Amphimedon queenslandica has been available for years at the Trace Archives but never assembled. The species was formerly called Reniera spp. and it is still carried under that name at JGI Genome. Consequently tblastn of contigs is not available without do-it-yourself assembly.
The Oakley group reported searching for sponge opsins but finding only "non-opsin, rhodopsin-class GPCR genes" from Amphimedon. Similarly, no opsins were located in the even earlier diverging placozoan Trichoplax, choanoflagellate Monosiga, and fungal genomes. This fits a picture of photoreceptor opsins first appearing subsequent to sponge in eumetazoa cnidarians. However these were not de novo genes but rather evolved out of the already-rich cauldron of GPCR gene copies.
Some later diverging species such as the model organism C. elegans lost all of their opsin genes, making them useless in Urbilateran ancestor reconstruction. This argues for much more intensive genomic sampling so as to sidestep the widespread problem of gene loss in model organisms.
-=-=-= coming real soon! -=-=-
- Ciona (tunicate) urochordate opsins
- Branchiostoma (amphioxus) cephalochordate opsins
- Stronglyocentrotus (sea urchin) echinoderm opsins
- Saccoglossus (acorn worm) hemichordate opsins
- Xenoturbella plus Convoluta no opsins yet
- Schmidtea (planaria) opsins
- Platynereis (polychaete) lophotrochozoan opsins
- Capitella (annelid) lophotrochozoan opsins
- Lottia and Aplysia lophotrochozoan opsins
- Nematostella (anemone) cnidarian opsins
- Hydra (hydra) cnidarian opsins