Opsins underground
Introduction
The genome article for the naked mole-rat (Heterocephalus glaber) appeared in Nature in October 2011. That paper made remarkable inroads into understanding slowed aging in this species, the longest-lived of all rodents. The mole-rat is thus an appropriate genomic model species to get at adaptations to aging processes.
As a hystricognath rodent (Euarchontoglires), mole-rat is only superficially related by convergent evolution to blind mole-rats (eg Spalax, murid rodent), lawn moles (Talpa: Laurasiathere insectivore), golden moles (Amblysomus: Afrotheres) or marsupial moles (Notoryctes). None of these species have comprehensive data sets that would allow comparison of opsin degeneration patterns but in some cases retinal anatomy has been studied.
Rodent lifespans range from a few years in mice and rats to nearly three decades in beavers, porcupines, and squirrels in addition to naked mole-rats. These species are in separate phylogenetic clades, implying slow aging (respectively fast-aging in other rodents?) has evolved on multiple occasions.
The mole-rat has quite an unusual life history, spending its life in dark hypoxic underground tunnels but in rare eusociality. The question arises as to what has happened to opsins and the many other components of the visual repertoire. Here it must be noted that "at lower temperatures, they bask in the shallow, more sun-warmed parts of their burrow systems" and that photos clearly show black eye spots implying melanin in the RPE (retinal pigmented epithelium).
This basking might also permit the photoisomerization step in the synthesis of vitamin D, though "intestinal calcium transport in mole-rats is independent of both genomic and non-genomic vitamin D" and vitamin D endrocrine receptors and hydroxylating enzymes are are present.
Photolyases, a class of flavo-enzymes involved in repair of UV-caused dna damage (thymine dimers), also utilize visible light in the reaction mechanism in bacteria. These enzymes are distantly homologous to mammalian CRY1 and CRY2 genes that entrain circadian rhythms perhaps via blue light reception in the retina but no longer repair dna. Light-driven magnetoreception has also been proposed in the subterranean mole-rat Cryptomys as well as human.
Opsin retention has been before in subterranean mammals, bats, dolphins, nocturnal primates and cave fish, with the outcome consistent with "use it or lose it" -- genes quickly degenerate by mutation as selective pressure for their maintenence ceases. Because the same genes are often used in both vision and hearing, utility in one system could maintain a given gene in both. Subterranean animals often have greatly reduced hearing and vision.
The mole-rat, like most placental mammals, has genetic potential for two-color cone vision plus dim-light rhodopsin in addition to five other opsins (peropsin, neuropsin, melanopsin, encephalopsin and rgropsin whose anatomical sites of expression (in the absence of mole-rat data) are likely the same as in mouse. The functions of these other opsins -- with the exception of rhabdomeric melanopsin -- are poorly understood but must be important because of their long history of conservation. The types and distribution of cone cells -- notably cone cell sharing -- in the mole-rat retina has not yet been determined.
The genome of last common ancestor of living amniotes contained a remarkable 21 opsins (defined as full length paralogs to rhodopsin with covalently bound retinoid at lysine-296). However by the time of earliest mammals, 9 of these opsins had already been lost; an additional two were lost in placental mammals. In terms of rod and cone opsins, mammals originally had 4-color vision along with colored oil droplet wavelength filters like lizards, turtles and birds still do.
Degeneration of the mammalian retina was the subject of a 1942 book by Gordon Walls. Although opsin were not sequenced until 50 years later, he anticipated their loss from detailed anatomical considerations. Walls proposed that mammals experienced a multi-million year bottleneck of deep forest nocturnality (rather than cave, marine or subterranean lifestyle) during their evolution and attributed opsin loss to loss of darwinian selection. Cone opsin loss has been previously studied in marine mammals (dolphin SWS1 opsin pseudogenized) and fruit bats (again SWS1 loss).
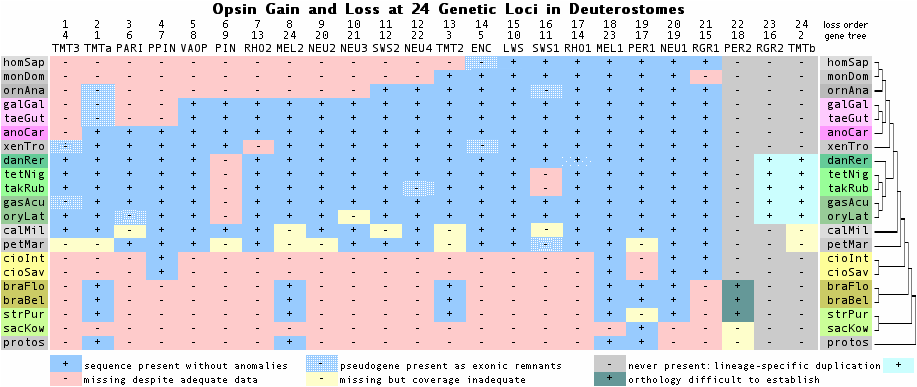
The figure above -- derived from exhaustive opsin gene family annotation in all available genome projects -- shows that the immediate rodent ancestor of naked mole-rat (NMR) had eight phylogenetically expected opsins, as mouse does today. The Nature article addresses the opsin portfolio of mole-rat but constrained by article size limits and the many other genes deserving commentary.
According to the article,
"We further examined the molecular basis for poor visual function and small eyes in the NMR (naked mole-rat). Of the four vertebrate opsin genes (RHO, OPN1LW, OPN1MW and OPN1SW), two (OPN1LW and OPN1MW) were missing; this distinguishes the NMR from other rodents with dichromatic color vision, such as mice, rats and guinea pigs. However, the NMR has intact RHO (rhodopsin) and OPN4 (melanopsin), supporting the presence of rod-dominated retina and the capacity to distinguish light/dark cues."
"Of about 200 genes associated with visual perception (GO:0007601) in humans and mice, almost 10% were inactivated or missing. These mammalian genes participate in crystallin formation, phototransduction in the retina, retinal development, dark adaptation, night blindness and colour vision.... Inactivation of CRYBA4, a microphthalmia-related gene, may be associated with the small-sized eyes, whereas inactivation of CRYBA4 and CRYBB3 and a NMR-specific mutation in CRYGS may be associated with abnormal eye morphology. Thus, while some genes responsible for vision are preserved, poor visual function may be explained by deterioration of genes coding for various critical components of the visual system."
Gene Protein Description Inactivation event Time of gene loss OPN1MW MWS mid wavelength complete loss detected with synteny in NMR stem lineage OPN1LW LWS long wavelength complete loss detected with synteny in NMR stem lineage -------- OPN1SW SWS1 short wavelength none retained to present EHB08658 RHO RHO1 dim rhodopsin none retained to present EHB14304 OPN3 ENC encephalopsin none retained to present EHB09988 OPN4 MEL1 melanopsin none retained to present RRH PER peropsin none retained to present OPN5 NEUR1 neuropsin none retained to present RGR RGR rgropsin none retained to present
The first two lines of the article summary table are reproduced above. The other lines are derived here using Blat of mouse opsin repertoire queries. Note the first line OPN1MW is an outright error as this gene is well-known to be a tandem duplication restricted to higher primates with no applicability to other mammals. These duplicates never existed in any ancestor of mole-rat and so was not lost in mole-rat. The genome assembly has good quality in this region; no tandem duplication of opsins specific to the mole-rat lineage occurs.
The second line is also in error. The terminal exon of mole-rat OPN1LW (LWS) is readily recovered by Blat of mouse query against mole-rat assembly. This gene fragment occurs in mole-rat in the expected syntenic location IRAK1 MECP2 OPN1LW TEX28 with conserved strand orientations. Because of assembly gaps, this exon could be either part of an intact gene or be all that is left after a large deletion, as discussed in depth below.
The article does not discuss OPN1SW, the short wavelength opsin lost in marine mammal and bat. This cone opsin is however present in mole-rat and little diverged from available rodent orthologs. This suggests functionality, apart from a six residue deletion in exon 4 associated with a frameshift in a run of six thymidines TTTTTT that would preserve reading frame if seven. This may represent inability of short read technology to precede past a compositionally simple region and allow read tiling rather than an actual deletion. No assembly gap is shown in the relevant region (JH170314:3551709-3551938). This region affects the lysine to which retinal is attached the region has to be carefully re-examined.
The other six opsins expected in mole-rat are readily located and appear intact apart from a small number of missing exons. The annotation provided at GenBank for encephalopsin (OPN3) is erroneous. As in all genome assemblies, that of mole-rat has incomplete coverage. Here eight genes were expected and eight can be detected. However only 11 of the 50 expected exons are represented (78% exonic coverage). However if a deletion has taken out 5 exons of OPN1LW (LWS) then coverage is better -- 6 out of 50 missing of 88%.
This article will use standard protein acronyms when assigned gene names have little mneumonic value, do not follow HGNC rules for paralog nomenclature, and do not exist at all for ancestral opsins still present in other amniotes. The table above provides nomenclatural correspondences.
Evolution of individual mole-rat photoreceptors
The eight opsins of mole-rat may be subtle or not-so-subtle pseuodgenes or at least evolving very rapidly away from conventional opsins under reduced selection attributable to the subterranean low or no light condition. To understand that, each of the mole-rate opsins is aligned with all the available rodent orthologs, with lagomorph opsins as outgroup. In some cases, large numbers of primate and laurasiathere opsins are included to provide additional context.
The alignments are shown as differences with respect to mole rat (top sequence). When assembly or deletional gaps occur in mote-rat opsins, predicted consensus sequences are shown instead (lower case). Deeply conserved residues that differ in mole-rat are quite evident as columns of conserved residues in these other species. These residues, and the number of them, are key indicators of the extent of anomalous evolution.
Rhodopsin, melanopsin and non-imaging opsins
Rhodopsin (RHO1 RHO): The alignment of mole-rat with a few of the thousand available orthologs shows no anomalies (subtle changes suggesting that the mole-rat gene is in functional decline). There is no known precedent for rodopsin loss in any vertebrate, even nocturnal cave bats, though a close paralog RHO2 persisted to contemporary lamprey through amniotes with loss only in stem mammalians.
The primary issue is whether mole-rat rhodopsin has adapted from an already low-light opsin in ancestral non-subterranean hystricognath to functional at even lower light levels. Bright sunlight might enter the top of a burrow and extend a long ways into tunnels. Even dim light might entrain circadian rithyms; fluctuations in contrast might indicate that a predator has entered the system.
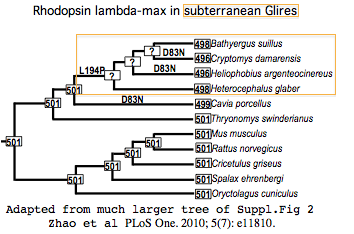
Rhodopsin adsorbs light maximally at 501 nm in most mammals and ancestral reconstructions. The shift in peak wavelength adsorption due to substitutions at the relevent amino acid positions has been reliably calculable for decades. These effects were specifically considered in a recent sweeping study of rhodopsins in low-light taxa.
The 13 tuning residues for lambda-max are shown with asterisks above the mole-rat sequence. Zhao et al calculated the effect of L194P in Heterocephalus (their independently determined rhodopsin sequence is identical to genomic) as lowering peak adsorption to 498 nm. The double variation D83N and L194P occurs in tuning residues of Cryptomys damarensis and Heliophobius argenteocinereus, reducing lambda-max there to 496 nm. The largest shift known is 17 nm.
Thus all three subterranean species of mole-rats have slightly reduced wavelengths at which their rhodopsins are most sensitive. While photobehavior -- notably burrow plugging -- has been directly studied, there is no experimental spectral data on tunnel light, so it remains unknown whether the dim light there is significantly stronger at these shorter wavelengths.
Note tunnels are very different optically from a clear water column where longer wavelength light is preferentially lost and from turbid water where particle scattering dominates. Given rhodopsin can already signal single photons, rod sensitivity has few prospects for further improvement. Nocturnal such as aye-aye have evolved enormous retinas for low-light imaging whereas mole-rats have little use for it.
In summary, there is little support for either adaptation or degeneration in mole-rat rhodopsin. With SWS1 and possibly LWS are retained in mole rat, rhodopsin is not likely to team up to provide 2-3 color vision as its subcutaneous eyes are distinctly vestigal (1.3 mm diameter). However cone cells amount to 10% of the mole-rat retina, far more than in mouse.
* * * *
RHO1_hetGla MNGTEGPNFYVPFSNITGMVRSPFEYPQYYLAEPWQFSMLAAYMFLLIVLGFPVNFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVICGFTTTLYTSMHGYFVFGATGCNMEGFFATLGGEIALWSLVVLAIERYMVVCKPMSNFRFGENHAIVGVAFTWVMALACAAPPL
RHO1_helArt ........V.............................M.................................N.......................P.............................V.................M..T..............
RHO1_cryDam ........V...................V..............I............................N.......................P..............................................VM.................
RHO1_batSui ........V.........F........................I.................................................I..P.............................V.................M...L.............
RHO1_thySwi .....A..V..................................I.................................FG.........L.......P....L........................V.................M.................
RHO1_cavPor ......E...I....A..V...................I......M.......I............................N....LG..........N......P....L........................V.................M..V...I..........
RHO1_musMus ...............V..V......Q...........................I.................................FG.........L.......P....L........................V.................M..V...I..........
RHO1_ratNor ..................V......Q...........................I.................................FG.........L.......P....L...........G............V.................M.................
RHO1_criGri ...............A..V..................................I.................................FG.........L.......P....L........................V.................M..V...I..........
RHO1_nanEhr ...............G..V......Q...........................I..........................L.......G.....F...L.......P....L........................V.................T.............V...
RHO1_oryCun .......D..I.M..Q..V..................................I.................................LG.........L.......P....V........................V.................M......I..........
* ** * * * * * *
RHO1_hetGla AGWSRYIPEGMQCSCGIDYYTPKPEINNESFVIYMFVVHFTIPLVIIFFCYGQLVFTVKEAAAQQQESATTQKAEKEVTRMVIIMVIAFLICWVPYASVAMYIFTHQGSDFGPIFMTIPAFFAKSSSIYNPVIYIMMNKQFRNCMLTTICCGKSPLGDDEASTTASKTETSQ
RHO1_helArt ....................................................................................................V..................S.................................N.......Y....
RHO1_cryDam ....................................................................................................V....................................................N............
RHO1_batSui ....................................................................................................V....................................................N............
RHO1_cavPor V...............V....L.S.V.H..........Y....MI......E................................................A........N.......V...................................N..........V.......
RHO1_thySwi .....................L...V.................MI................................................................N.......L.......A...........................N............
RHO1_musMus V....................L...V.................MIV.........................................F.....L......F........N.......L..................L...........L....N.....D..A.........
RHO1_ratNor V....................L...V.................MIV.........................................F.....L...............N.......L......TA.....I................L....N........A.........
RHO1_criGri V...............V....L...V..................IV.....................................L..VF.........G..F........N.......L..............................L....NI.......A.........
RHO1_nanEhr V....................L...V.................MI................................................................N.......L.......A......................L....NL...E...P.........
RHO1_oryCun V....................L...V..................I.......................................................F........N...........................................N........A.........
Melanopsin (MEL1 OPN4): This gene consists of 10 exons, very unusual for the opsin family. Because splice donors and acceptors are under selective pressure for both preservation of splicing and the role if any of the amino acids at the junctions, they evolve under different circumstances. In the mouse/rat/hamster clade, a synapomorphic deletion of one residue occurs near the end of the 9th exon. An alternative splice donor also occurs a few residues back, giving rise to the 'short form' of melanopsin in this group. (The same acceptor is used.) Many gene compilations err in providing this short form as the standard gene model -- the long form is ancestral and continues to be used even in this narrow clade.
Melanopsin has great natural variability in its first and ninth exons, so the variability seen in the difference alignment relative to mole-rat is *not* indicative of pseudogenization. -- the same would occur for any species in lead position. These regions lie not only outside the seven transmembrane region of this GPCR but also beyond the region of interaction with transducins with the cytoplasmic tail. Melanopsin, the sole rhabdomeric opsin in tetrapods, may be losing distal functions that it evolved and retained in invertebrates. Note the 3 residue deletion in the ninth exon -- this may be a valuable synapomorphy for refining hystricognath phylogeny.
Mole-rat does have five oddities (shown in red) supportive of a very early stage of pseudogenization -- non-conservative amino acid substitutions at positions highly conserved in other rodents (and other mammals). However if more data were available for hystricognath rodents, these might more widespread, indeed adaptive. The two critical regions for signalling and retinal binding, shown in blue, are intact. On balance, mole-rat melanopsin is very likely to be fully functional.
_ _ _
MEL1_hetGla MDTPSGPRVSPDPAQEPSFLATLSAP-GRGDGTPSSLSTPGQLPPGNPT AAGTQAAVRVPFSTADVPAHVHYTLGAVILLVGLTGMLGNLTVIYTFFR SRGLRTPANMFVINLAVSDFLMSFTQAPVFFTSSLYKRWLFGEA GCEFYAFCGALFGITS
MEL1_cavPor MEP.SG.T.GS.VS...P...S...D.L...F..AP....S.. V..A...AWI..P.V.....A..........................C. ...........I................................ ................
MEL1_musMus ..S......LSSLT.D...TTSPAL .QGIWNGTQNV.VRA..LSVS.. TSAH...AW...P.V...D.A.....T....................C. N..........I...........V.......A.....K.....T ..........V.....
MEL1_ratNor .NS..ES..PSSLT.D...T.SPALL.QGIWNSTQNI.VRV..LSVS.. TP.L...AW...P.V...D.A.....T....................C. N.........LI...................A.....K.....T ..K.......V...V.
MEL1_criGri ..S....TGP.GLT.G...M.STAL .QGHWNSTQKV..RV..LSMS.. .S.PE..AW...P.V...D.A.....T....................C. N..........I...................A.....K.....T ..........V.....
MEL1_phoSun ..S.P..TAP.GLT.G...M.STTL .HSHWNSTQKV..RA..LAVS.. .S.PE..AW...P.V...D.A..I..T....................C. ..S.......LI...................A.....K.....T ..........VL....
MEL1_nanEhr .NS......P.GL..K...MV.PVL..NQWISFQKNV.VGI....ASA. .T.A...SW...P.V...V.A.....T..............I.....C. ......R....TV..................A............ ..........VS....
MEL1_dipOrd .NP..R..I..N.T.....A..PAS....W.S.Q.NV...S....IST. T..........I...................A...H.Q...... ................
MEL1_ictTri .N.T..SD ..G.....TLG..SAP..SMW.S.Q..V.SLA..L.AS.. .T.A.S.A.I..P.V...D.A.....T.........LM.........C. ..S........I................I..A............ ..........V...S.
MEL1_oryCun .NS.W.S..P.G.....R S.AAP..S.W..SE..I.S....T..S.. .P.A.E.AW...P.V...D.A.....T....................C. ..S........I...................A...........T ..............S.
MEL1_ochPri ..P...S..LAS.T....SNSSSA..AN.W.SSQN.P.SL...SSSS.. VLVV.D ..P.V...D.A.....S....................C. ...........I...................A.......I...T ..............S.
_ _
MEL1_hetGla MITLTAITLDRYLVITRPLATIGLASKRQAALVLLGIWLYALAWSLPPFFGW SAYVPEGLLTSCSWDYMTFTPSVRSYTMLLFCFVFFLPLLIIIYCYVFIFRAIRETGR AFESCGESPSRRRQWQRLRSEWKMAKIALLVILLFVLSWAPYSTVALV
MEL1_cavPor .......................V............V............... ................V.......A...............V.....I........... .........R.................................A....
MEL1_musMus .......AM..............RG...RT......V............... .....................Q..A..................F..I........... .C.G.....L........Q.......V..I..................
MEL1_ratNor .......AM..............MR...RT......V............... ................V....L..A..................F..I........... .C.G.....L........Q.......V..I..................
MEL1_criGri .......A...............MG...RT.F....V............... .....................Q..A..................G..I........... .C.G.S...QQ....H..Q.......V..I..................
MEL1_phoSun .......A...............MG...RT...................... ................V....Q..A...............V..F..IS.......... .C.GWS...Q.....H..Q.......V..I..................
MEL1_nanEhr .T.....A...............V....RT......V............... ........................A..................F..I...K....... .C.G.....Q........QN......V.....F...............
MEL1_dipOrd ....M..A...............VT...RT.F....V.V............. ........................................V.....I.........R. .C.G.....QQ.Q.....Q.............................
MEL1_ictTri .......A...............M...KR..FF...V.F............. ........................A.....V........FL.....I........... .C.G.....R.......MQ....V...........IV......I....
MEL1_oryCun .......A............AV.MV..KR.G.....V...........L... ................V.......................V.V...I........... .CQGSH...Q.. ..R..Q.......V........L............
MEL1_ochPri .......A............AV.MV...RTG.....V...S..C....L... ........................A.....V........FL.....I........... .....L............
_ _ _ _
MEL1_hetGla AFAG HAHILTPSMSSVPAVIAKASAIHNPIIYAIAHPKY RMAIVQHLPCLGLLLGVSGQRSLPSLSYRSTHRSRLSSQASDLSWISGRRRQESLGSESEV GWTDTE---AWGSAQQASERPPCGTGLEGWEARAHPKAQEWEAETPGK TKGRLPSLDP*
MEL1_cavPor .... Y..M...Y.N....................T.... A ..........V.......HGR...R.......T.......................... ......---...A...G.GQS.YDLV..D.....P...RV.G.G.... ...Q.TC...
MEL1_musMus .... YS.....Y......................T.... .V..A.......V.........H...........T....S.........K........... ......TTA...A.....GQSF.SQN..DG.LK.SSSP.VQRSK..-. ...H.....L
MEL1_ratNor G... YS.....Y......................T.... .A..A.......V.........H...........T....S........QK........... ......TTA...A.....GQSF.SHD..DG.VK.PSSP..QKSK..-. ..RH.....R
MEL1_criGri .... YS.....Y......................T.... .A..A.......V.....S...H........Q..T....S........PK........... ...E..ATAV..T..P..GQSS.SQN.QDGAVK.PSSP.VQNSKA.-. A..Q.....L
MEL1_phoSun .... YS.....Y..................V...T.... .A..A.......V.....S..NR...........T....S.......APK........... ......ATAV..A..P..GQSS..QN..DGMVK.PSSP.VQNSKA.-. A..Q.....L
MEL1_nanEhr .... YS.....Y.N....................T.... .L..S.......V.I...S...H...........T..............K........... ......VTA...V..E..GWS.YRHS..DG.VK.S.SP.GQ..K.SR. ...Q....NL
MEL1_dipOrd .... Y......Y.N..........S.....V...T.... .V..A......RV.......HDH....F......T.T..T...................KM ......AAA...D...MTGQS..SQD..DG.VKVL.R..RK..K..K.
MEL1_ictTri .... Y..L...Y......................I.... ....S.N..F.KK.....S.HGR...........T.I.H..EV.................. ......AAA...A...MNGWSL..QI..DE..T.PSRS.GQ....FR. ...Q....CS
MEL1_oryCun .... Y..S.S.Y.N....................T.... .A..A.......V......RH.R...........T.......................... ......AAA...A.L.L.G.YL..Q...DG.IK.T.RR.GP.....R. ..RLR.C...
MEL1_ochPri .... ...S...Y.T....................T.... .V..A.......V.......H.R...........TV...T.................V... ..A...VAA.Y.ASPLV.G.YLY.QV..NE..K.A.RCHGPG....R.
Encephalopsin (ENC OPN3):
ENC_hetGla mysgnrsggqgywegggaagaegpapagtlspaplfspgayerlalllgslgllgvgnnllvlvlyykfqrlrtpthlflvnislgdllvslfgvtftfVSCLRNGWVWDAVGCVWDGFSSSLFGIVSITTLTVLAYERYIRVVHARVINFSWAWKAITYIWLYSLAWAGAPLLGWNRYILDVHGLSCTVDWKSKDANDSSFVL ENC_cavPor .......S........ .P.D..................................................S......A....S...G.............K................R..................................R..........................I..................... ENC_musMus ........D.....D. .....A.....R........T..........C.A.....G.....L..S..P............L................A..............A.....G....F.............................R..........................I...G.....R........... ENC_ratNor ..............D. .....A.....R........T..........C.A.....G.....L..S..P............L................A..............A.....G....F.............................R..............................G................. ENC_speTri ........S..S...D.S.....S..E.....T......TN......FR.V....A.S............GSAH.LTF...........M...............R....T.A..........................................R..............................G...E............. ENC_criGri ........L................A..............A.....G....F...................S.........R..........................I...G................. ENC_dipOrd ..........E...D.....S................A............A...........................L......S........................T.........R..............................T...R..........................I...G......A.......... ENC_oryCun ........E...............G.............ST..........I......S..................L........S.....V..................T............................................R..........................I...G........N........ ENC_hetGla FLFLGCLVVPMGVIAHCYGHILYSIRMLRCVEDLQTIQVMKILRYEKKVAKMCFLMVFIFLVCWMPYIVICFLVVNGYGHRVTPTVSVVSYLFAKSSTVYNPVIYTIMIRKFRRSLLQLLCFRLLRCQQPAKDLPAVESEMQIRPIVMSQKDGDRPKKKVTFNSSSIIFIITSDESLSVDDSDRTSGSKVDTIQVRPL ENC_cavPor ..........V...V..............G..............S.N...I......................L....R........I.....T...........VL............H.L................R..H........................................................ ENC_musMus ..........V.I...........V..............I.M..............A.V..T.......TR.........L......I.................IF.N...................R...N...A....H.....................................E....S.A....V...... ENC_ratNor ............I...........V..............I.M..............A.V..T.....V.TR.........L......I.................IF.....................R...N...A..........................................E....S.A....V...... ENC_speTri ..........V........................IF..I........L.....V...T..I.......V....A....Q.......I..N..............IF..............S...............GN........I.....E.............V..............N......A.V...... ENC_criGri ..........L.I.T.........V..............I....C.......S.A...V..T.......TR.........L......I..S...........I..IF.....................R...N...A...............R................................NA..ANV...... ENC_dipOrd ...I......V.I.........................II...Q....L.......ALT..M.......T......SH..L....I.I..H.L.....I......IF.....................R.......AG............................................V.S....A.V...... ENC_oryCun ..........V.............V..............I..............F...T..I.....V............L....L.I.....C....A...I..IF...............QP......P....T.G.................................A.....A...NEKA..P...V......
Neuropsin (NEUR1 OPN5):
NEUR1_hetGla MALNHTAPPQDERLPHYLQDEDPFVSKLSWEADLVAGFYLTIIGILSTCGNGYVLYMSSRRKKKLRPAEIMTINLAICDLGISVVGKPFTIISCFRHRWVFGWIGCRWYGWAGFFFGCGSLTTMTVVSLDRYLKICYLSYGVWLKRKHAYICLAAIWAYVSFWTTMPLVGLGDYAPE NEUR1_cavPor ..........N.H..R................................V........................................................................I....................................................... NEUR1_musMus .......L..........R.....A.......................F...........................V..................C.......F.................I...A............................V....A................. NEUR1_ratNor .......L..........R.....A.......................F...........................V..................C.......F.................I...A............................V....A................. NEUR1_speTri .......L....H.....R.....A.......................F...........................V..................C.........................I...A.......................F....V....A................. NEUR1_dipOrd ..F....GT.GQG.....PE....T........I..............F...........................V..................C.........................I...A............................V....A..............V.. NEUR1_oryCun .......L....H.....REG...A.......................F...........................V..................C.........................I...A....................R.......L....A................. NEUR1_ochPri ....D..L....H....FR.G...A.......................F...........................V..................C...........L....D........I...A........ ......R.......V....A................. NEUR1_hetGla PFGTSCTLDWWLAQASARGQVFILNILFFCLLLPMATIVFSYAKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTKVAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYRFACCQNGGLKATKKKSLEDFRLHTVTTVRKSSAVLEIHQEV NEUR1_cavPor .................G..I...H.........T.M.....V............I.............................................................................S.......A....................D... ......... NEUR1_musMus ................GG......S.........T.V.................................V.....................................................................A...RG.............................. NEUR1_ratNor ................GG......S.........T.V.................................V................................N....................................T...R................A...........P.. NEUR1_speTri ................VG..... ..........T.V.E...V..........E.............. .......................................S...................K.....T....................A.......V...... NEUR1_dipOrd ................LA................TSV.....V....................P........... NEUR1_oryCun ................VG................T.V.....V...........................................................................................K.S..RTS.................................. NEUR1_ochPri ................VG................T.V.....V.....................G.....................................................................K.S..RT....Q..............................
Peropsin (PER RRH):
PER_hetGla MLRNSFGNGSDSKSEDGSVFSRMEHSIVAAWLLLAGLISILSNIIVLGIFVKYKELRTPTNAIIMNLALTDIGVSSIGYPMSAASYLHGSWKFGYPGCQVYAGLNIFFGMASIGLLTVIAVDRYLTICRPDIGRRLTSTSYVSMILGAWINGLFWALMPIIGWASYAPD PER_cavPor ...H.L..S....N.......QT..N....Y.I.................I..................................D.........A......................V................M..H...G.......................... PER_musMus ..SEASD.S.G.R.. ......T...VI..Y.IV..IT.....VV.....I...........V.I...F................D........HA...I..........V.......V.M......SC..V...M.TNT.L........................... PER_ratNor ...DALD.S.G.G.. ....TKS....I..Y.IV..I.............I...........V.I...F................D........HA..............V.......V.L......SC..V...M.GNT.L..V...............V........ PER_dipOrd ....NV..S.G.RN.......QT..N...TY.IT..V......L......I.............I....................D.Y.R.....A...I..................V.I.......H.....GM.TRT..T.......................... PER_oryCun ....NLS.S..F.H.......QT..N...TY.I...M......L......I.............I...F................D.........A...I..................V.M.......H..V...M.TRT.LGL.....V..........A........ PER_ochPri ...HNL..S.EA.V.A.....QT..N....Y.I...M......L......I.............I...F................D.........A...I..................V.M.......Q......M.THT.FG.................V........ PER_hetGla PTGATCTINWRKNDTSFVSYTMTVIAVNFTGPLAVMFYCYFHVTWSIKHHATGNCPTFPNRDWSDQVDVTKMSVVMILMFLVAWSPYSIVCLWASFGDPKKIPPSMAIIAPLFAKSSTFYNPCIYVIANKKFRRAMCTMFRCQTHQAMPETSALPMGASQSPLASG PER_cavPor ..............VA..........I..IV.........L.I.RA.RR.VA.DR.PNLSG......................................RR.S.................................FA..Q......V.VA.I...D.........RI PER_musMus ...........N..........M..V...IV..T......Y..SR.LRLY.ASD.TAHLH...A..A.......I......L............C..N............................A.H....K..LA..K..P.L.V..P.T...DMP..S..PVRI PER_ratNor ......................M..V...IV..T......Y..SQ.MRLS.AS..T.HL....AH.A.......M......L......V.....C..N.......L....................A......K..FA.LK..P......P.T.A..VPH....PARI PER_dipOrd ............... .............................E...P.....................V.........LA.LK........V..I...DV..N.....RI PER_oryCun ...................F..A...I..VV..T......Y...Q...Q.RASD.TEYL...............I..F..........................A.....................A...R.....FA..K........V..V...DV..N..P..II PER_ochPri ..............K....F..A..M...VV..T......YY..Q.....TASD.TKSL...............I........................Q..........................A...RS....FA..K..IP..K.V..LS.RDV.....S..RT
Rgropsin (RGR):
RGR_hetGla MAASGALPTGFGELEVLAVGTMLLVEALCGLSLNGLTIVSFCKIPELWTPRHLLVLSLALADSGLSLNTLIAAVSSLLRRWPYGSGGCQTHGFQGFVTALASISGSAAIAWGHHQHYCTGSQLAWSTAIRLVLFVWLSSAFWAALP RGR_cavPor ..T.E...A............V..L.G..........V...W.S.A.R..N.................A.V..G.....H.. ...H..AL.....T........T..LS..R..QC..RGR.T....VP................ RGR_musMus ...TR...A.L..........V..M...S.I.......F....T.D.R..SN..........T.I...A.V...........H..E...V......A......C....V...RYH.....R....D...P......M......S.. RGR_ratNor .T.TR...A.........I.IV..M...S.I.......F....T.D.R..SN..........T.I...A.V...........H..E..RV......A......C........RYH.....R....D...P.............S.. RGR_speTri ..ETA...A............V......S.........F....T...R..N...L....V....I...A....I...........D...A..........S..C........RYH..........N...P.........T...... RGR_dipOrd ..T..D...........T...V......S.....T...F....T...R..I...D....V....I...A....I...EWH....LE...A......................RCH.H....L.G.D..VS..I............. RGR_oryCun ..EP.T..P..E.........V......S.........F....T......S........V....I...A................D...A......A......C........RYH..........N..VL.........V...... RGR_ochPri ..EP.T..P..E.........V......T.....S...F...TS...R..S.............V...A.A..TA..........D...A......A......C........RYH..........N..VL.........V...... RGR_hetGla LLGWGHYDYEPLGTCCTLDYSKRDRNSTSFLFTMGFFSFLIPLFITFTSYQLMEQKLARSSRLQVNTSLPARTLMLCWGPYVFLHLYAVIADASSLSPKLQMVPALIAKTVPTINAINYALGSKMGSRRPWQCLSLQRSKRD RGR_cavPor .....R...............TG...F.......A..N..V.....V..C....RH.........SVRQ.....L...S..AL.Y....L...HT...R............... .S.. .G...S.EM....Q.... RGR_musMus .M.........V.........RG...FI......A..N..V.....H...RF....FS..GH.P...T..G.M.L.G....AL.Y...A...V.FI..............M..........HRE.VC.GT.....P.K..K.RTQ RGR_ratNor .M.........V.........RG...FI......A..N..V.....H...RF.....S..GH.....T..G.M.L.G....AL.Y...AV..V.FI..............M..........R.E.VC.GT...R.A.K..Q.RTQ RGR_speTri .....................RG...FI......A..N.FV.....L...R.........GH.. ... RGR_dipOrd .......N.............RG...F.......A..N..V.....L......K..F...G......T..T...L.G....AL.YF..A.M.VN.I.............M...V.......CNELLCGGFSLG.LP.KG.Q.RTQ RGR_oryCun .....................RG...FI...I..A..N..M.....L...S......SK.G......T..G...LF.....AV.Y.C.AV..M..ITL...............V..V......EVIR.GI....LP...V.GRAQ RGR_ochPri .....................RG...FI...V..A..N..M....ML...S.......K.G......T......LF.....AI.C.C.TVM.M.TV....L........A...V.........EVIR.GI....LP...V..RAQ
Does mole-rat have OPN1LW (LWS) or just a residual exon?
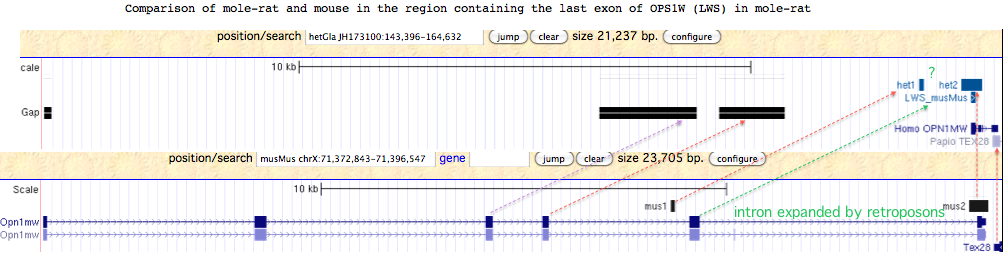
Since the only detectable OPN1LW exon of mole-rat lies in a region with several large assembly gaps corresponding to the missing exon placement in mouse genome, mole-rat may have a complete functional copy of OPN1LW. Alternatively, the observed genomic compression of this region relative to mouse suggests a large deletion occurred in mole-rat.
Although exons 3 and 4 of mole-rat OPN1LW may lie in assembly gaps, it is not clear why the penultimate exon 5 cannot be recovered by blastx from the region indicated by the green arrow in the figure below. Rapid lineage-specific gain and loss of retroposons can cause intronic and inter-genic regions to diverge very rapidly. Here, the sole correspondences of RepeatMasked sequence between mole-rat and mouse are indicted by the two small regions denoted het1/2 and mus1/2.
Conservation of the amino acid sequence of the sole exon present is high; only very recent pseudogenization is compatible with mostly mild substitutions and no internal stop codons or frameshifts. However the proline replacing arginine -- if the sequence here is correct -- is assuredly disabling in any imaging opsin in any metazoan. This arginine is slightly distal to the retinal binding motif and critical to opsin transducin signalling even in non-opsin GPCR.
OPN1LW LWS_hetGla FPNCILQLFGKKVEDSLELSSASKTEASSVSSVSPA* last mole-rat exon aligned to mouse
F.NCIL.LFGKKV.DS.ELSS.SKTE.SSVSSVSPA* 6 differance out of 36 residues
OPN1LW LWS_musMus FRNCILHLFGKKVDDSSELSSTSKTEVSSVSSVSPA*
The alignment below shows that exon 6 of mole-rat OPN1LW (LWS) has three unusual substitutions but overall is highly conserved as are all mammalian LWS sequences. The proline substituted for arginine at position two is a radical substitution both in terms of physical attributes and site-specific conservation. At the dna level, this represents a CpG hotspot mutation, CGA to CCA. Glutamate for aspartate is a mild substitution in many proteins but not at this site as it is tolerated only in guinea pig and rabbit. It is a transversion GAT to GAG here. Leucine for serine is again an extreme change in amino acid properties at a site not tolerating variation. It arose here as a CTT to CTC transversion in the split codon box of these six-codon amino acids. This region of the LWS protein constitutes the cytoplasmic tail with the usual seven-transmembrane GPCR structure.
The genomic sequence here represents a single male individual. The fragment-only haplotype and these substitutions may not be representative of the overall mole-rat population. Since mole-rats have standard placental XY sex determination and this gene always lies on mammalian chromosome X, males cannot have a compensatory normal copy. However male color blindness is common in other species including human.
In the scenario of balanced polymorphism at this locus, females of a species might attain a measure of trichromatic vision whereas males could not. Should the balancing polymorphism be this deletional pseudogene, the male population would be an equal mixture of functioning and non-functioning at this locus whereas only a quarter of females would. This conceivably could correlate with mole-rats living in large colonies with a single breeding queen.
Sole exon of mole-rat OPN1LW (LWS) Difference alignment relative to other mammals Explanation of genus species codes
LWS_hetGla FPNCILQLFGKKVEDSLELSSASKTEASSVSSVSPA LWS_hetGla FPNCILQLFGKKVEDSLELSSASKTEASSVSSVSPA Heterocephalus glaber (naked_molerat)
LWS_musMus FRNCILHLFGKKVDDSSELSSTSKTEVSSVSSVSPA LWS_musMus .R....H......D..S....T....V......... Mus musculus (mouse)
LWS_ratNor FRNCILQLFGKKVDDSSELSSTSKTEVSSVSSVSPA LWS_ratNor .R...........D..S....T....V......... Rattus norvegicus (rat)
LWS_nanEhr FQNCILQLFGKKVDDSSELSSTSKTEASSVSSVSPA LWS_nanEhr .Q...........D..S....T.............. Nannospalax ehrenbergi (mole_rat)
LWS_speTri FRNCILQLFGKKVDDTSELSSASRTEASSVSSVSPA LWS_speTri .R...........D.TS......R............ Spermophilus tridecemlineatus (squirrel)
LWS_cavPor FRNCILQLFGKKVEDSSELSSTSRTEASSVSSVSPA LWS_cavPor .R..............S....T.R............ Cavia porcellus (guinea_pig)
LWS_oryCun FRNCILQLFGKKVEDSSELSSASRTEASSVSSVSPA LWS_oryCun .R..............S......R............ Oryctolagus cuniculus (rabbit)
LWS_homSap FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA LWS_homSap .R...........D.GS.........V......... Homo sapiens (human)
LWS_panTro FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA LWS_panTro .R...........D.GS.........V......... Pan troglodytes (chimp)
LWS_gorGor FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA LWS_gorGor .R...........D.GS.........V......... Gorilla gorilla (gorilla)
LWS_ponPyg FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA LWS_ponPyg .R...........D.GS.........V......... Pongo pygmaeus (orang_sumatran)
LWS_nomLeu FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA LWS_nomLeu .R...........D.GS.........V......... Nomascus leucogenys (gibbon)
LWS_macMul FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA LWS_macMul .R...........D.GS.........V......... Macaca mulatta (rhesus)
LWS_macFas FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA LWS_macFas .R...........D.GS.........V......... Macaca fascicularis (macaque)
LWS_papHam FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA LWS_papHam .R...........D.GS.........V......... Papio hamadras (baboon)
LWS_calJac FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA LWS_calJac .R...........D.GS.........V......... Callithrix jacchus (marmoset)
LWS_cebCap FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA LWS_cebCap .R...........D.GS.........V......... Cebus capucinus (monkey)
LWS_ateJef FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA LWS_ateJef .R...........D.GS.........V......... Ateles geoffroyi (monkey)
LWS_otoGar FRNCILQLFGKKVDDSSELSSTSKTEVSSVSSVSPA LWS_otoGar .R...........D..S....T....V......... Otolemur garnettii (bushbaby)
LWS_otoCra FRNCILQLFGKKVDDSSELSSTSKTEVSSVSSVSPA LWS_otoCra .R...........D..S....T....V......... Otolemur crassicaudatus (bushbaby)full
LWS_micMur FRNCILQLFGKKVDDGSELSSTSKTEVSSVSSVSPA LWS_micMur .R...........D.GS....T....V......... Microcebus murinus (mouse_lemur)
LWS_tupBel FRNCILQLFGKKVDDSSELSSASRTEASSVSSVSPA LWS_tupBel .R...........D..S......R............ Tupaia belangeri (tree_shrew)full
LWS_canFam FRNCILQLFGKKVDDSSELSSASRTEASSVSSVSPA LWS_canFam .R...........D..S......R............ Canis familiaris (dog)
LWS_phoVit FRNCILQLFGKKVDDSSELSSASKTEASSVSSVSPA LWS_phoVit .R...........D..S................... Phoca vitulina (seal)
LWS_felCat FRNCIMQLFGKKVDDGSELSSASRTEASSVSSVSPA LWS_felCat .R...M.......D.GS......R............ Felis catus (cat)
LWS_bosTau FRNCILQLFGKKVDDSSELSSVSKTEASSVSSVSPA LWS_bosTau .R...........D..S....V.............. Bos taurus (cow)
LWS_turTru FRNCILQLFGKKVDDSSELSSVSKTEASSVSSVSPA LWS_turTru .R...........D..S....V.............. Tursiops truncatus (dolphin)
LWS_delDel FRNCILQLFGKKVDDSSELSSVSKTEASSVSSVSPA LWS_delDel .R...........D..S....V.............. Delphinus delphis (dolphin)
LWS_gloMel FRNCILQLFGKKVDDSSELSSVSKTEASSVSSVTPA LWS_gloMel .R...........D..S....V...........T.. Globicephala melas (pilot_whale)
LWS_susScr FRNCILQLFGKKVDDSSELSSVSKTEASSVSSVSPA LWS_susScr .R...........D..S....V.............. Sus scrofa (pig)
LWS_equCab FRNCILQLFGKKVDDSSELSSVSKTEASSVSSVSPA LWS_equCab .R...........D..S....V.............. Equus caballus (horse)
LWS_myoLuc FRNCILQLFGKRVDDSSELSSTSRTEASSVSSVSPA LWS_myoLuc .R.........R.D..S....T.R............ Myotis lucifugus (brown
LWS_pteVam FRNCILQLFGKKVDDSSELSSSSKTEASSVSSVSPA LWS_pteVam .R...........D..S....S.............. Pteropus vampyrus (flying_fox)
LWS_loxAfr FRNCILQLFGKKVDDSSELSSASKTEASSVSSVSPA LWS_loxAfr .R...........D..S................... Loxodonta africana (elephant)
LWS_triMan FRNCILQLFGKKVDDGSELSSASKTEASSVSSVSPA LWS_triMan .R...........D.GS................... Trichechus manatus (manatee)
LWS_echTel FRNCILQLFGKKVDDSSELSSTSRTEASSVSSVSPA LWS_echTel .R...........D..S....T.R............ Echinops telfairi (tenrec)
LWS_proCap FRNCILQLFGKKVDDSSELSSTSKMEASSASSVSPA LWS_proCap .R...........D..S....T..M....A...... Procavia capensis (hyrax)
LWS_monDom FRTCILQLFGKKVDDGSEVSSTSKTEGSSVSSVAPA LWS_monDom .RT..........D.GS.V..T....G......A.. Monodelphis domesticus (opossum)
LWS_didAur FRTCILQLFGKKVDDGSEVSSTSKTEVSSVSSVAPA LWS_didAur .RT..........D.GS.V..T....V......A.. Didelphis aurita (big-eared
LWS_tarRos FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA LWS_tarRos .RT..........D.GS.V..T.R..V......A.. Tarsipes rostratus (honey
LWS_macEug FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA LWS_macEug .RT..........D.GS.V..T.R..V......A.. Macropus eugenii (wallaby)
LWS_setBra FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVPA- LWS_setBra .RT..........D.GS.V..T.R..V......PA- Setonix brachyurus (quokka)
LWS_cerCon FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA LWS_cerCon .RT..........D.GS.V..T.R..V......A.. Cercartetus concinnus (pygmy
LWS_smiCra FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA LWS_smiCra .RT..........D.GS.V..T.R..V......A.. Sminthopsis crassicaudata (dunnart)
LWS_myrFas FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA LWS_myrFas .RT..........D.GS.V..T.R..V......A.. Myrmecobius fasciatus (numbat)
LWS_isoObe FRTCILQLFGKKVDDGSEVSGTSRTEVSSVSSVAPA LWS_isoObe .RT..........D.GS.V.GT.R..V......A.. Isoodon obesulus (bandicoot)
LWS_ornAna FRNCIMQLFGKKVDDGSELSSTSRTEVSSVSSVSPA LWS_ornAna .R...M.......D.GS....T.R..V......... Ornithorhynchus anatinus (platypus)
LWS_tacAcu FRTCILQLFGKKVDDGSEVSSTSKTEVSSVSSVAPA LWS_tacAcu .RT..........D.GS.V..T....V......A.. Tachyglossus aculeatus (echidna)
Within the broader context of imaging opsin comparative genomics, the glutamate/aspartate substitution is very common. While not without effects on interaction of the cytoplasmic tail with transducins, this change in mole-rat LWS is not likely to be disabling. The leucine/serine substitution is not located at a site important overall to imaging opsins; the reason it is invariant from lamprey to mouse/human in LWS is not known. The loss of arginine at 2nd position is completely disabling in any opsin (indeed any GCPR) because it is critical to the signalling mechanism.
P E L
LWS_hetGla FPNCILQL--FGKKV---EDSLELSSASKTEAS --SVSSVSPA
LWS_musMus KSATIYNPIIYVFMNRQFR NCILHL--FGKKV---DDSSELSSTSKTEVS --SVSSVSPA
LWS_homSap KSATIYNPVIYVFMNRQFR NCILQL--FGKKV---DDGSELSSASKTEVS --SVSSVSPA
LWS_monDom KSATIYNPIIYVFMNRQFR TCILQL--FGKKV---DDGSEVSSTSRTEVS --SVSSVAPA
LWS_ornAna KSATIYNPIIYVFMNRQFR NCIMQL--FGKKV---DDGSELSSTSRTEVS --SVSSVSPA
LWS_galGal KSATIYNPIIYVFMNRQFR NCILQL--FGKKV---DDGSEV-STSRTEVS SVSNSSVSPA
LWS_anoCar KSATIYNPIIYVFMNRQFR NCIMQL--FGKKV---DDGSELSSTSRTEVS SVSNSSVSPA
LWS_xenTro KSATIYNPIIYVFMNRQFR NCIYQL--FGKKV---DDGSEVSSTSRTEVS SVSNSSVSPA
LWS_neoFor KSATIYNPIIYVFMNRQFR NCIYQL--LGKKV---DDGSELSSTSKTEVS SVSNSSVSPA
LWS_takRub KSATIYNPVIYVFMNRQFR VCIMKL--FGKEV---DDGSEV-STSKTEVS -----SVAPA
LWS_gasAcu KSATIYNPVIYVFMNRQFR SCIMQL--FGKEV---DDGSEV-STSKTEVS -----SVAPA
LWS1_calMi KSSTIYNPIIYVFMNRQFR NCILQL--FGKKV---DDGSELSSTSKTDVS SVSNSSVSPA
LWS2_calMi KSSTIYNPIIYVFMNRQFR NCILQL--FGKKV---DDGSELSSTSKTDVS SVSNSSVSPA
LWS_petMar KGATIYNPIIYVFMNRQFR NCILQL--FGKKV---DDGSEVSSSSRTEVS SVSNSSVSPA
LWS_letJap KSATIYNPVIYVFMNRQFR NCIMQL--FGKKV---DDGSEVSSASRTEVS SVSNSSISPA
LWS_geoAus KSATIYNPIIYVFMNRQFR NCIMQL--FGKKV---DDGSEVSSSARTEVS SVSNSSVSPA
P E L
SWS1_hetGl KSSCVYNPIIYCFMNKQFR ACIMEL-VCRKSMA--DESDMSSS-QKTEVS AVSSSKVGPN
SWS1_musMu KSSCVYNPIIYCFMNKQFR ACILEM-VCRKPMA--DESDVSGS-QKTEVS TVSSSKVGPH
SWS1_homSa KSACIYNPIIYCFMNKQFQ ACIMKM-VCGKAMT--DESDTCSS-QKTEVS TVSSTQVGPN
SWS1_monDo KSACVYNPIIYCFMNKQFH ACIMEM-VCRKPMT--DDSDVSSS-QKTEVS AVSSSQVGPT
SWS1_smiCr KSACVYNPIIYCFMNKQFH ACIMEM-ICKKPMT--DDSETTSS-QKTEVS TVSSSQVGPS
SWS1_tarRo KSACVYNPIVYWFMNKQFH ACIMEM-VCRKPMT--DDSEISSS-QKTEVS TVSSSQVGPS
SWS1_taeGu KSSCVYNPIIYCFMNKQFR ACIMET-VCGRPMT--DDSEVSSSAQRTEVS SVSSSQVGPS
SWS1_anoCa KSSCVYNPIIYCFMNKQFR ACILET-VCGKPMS--DESDVSSSAQKTEVS SVSSSQVSPS
SWS1_utaSt KSACVYNPIIYCFMNKQFR ACIMET-VCGKPMT--DESDVSSSAQKTEVS SVSSSQVSPS
SWS1_neoFo KSSFVYNPIIYCFMNKQFR ACIMQT-VFGKPMT--DDSDISSSG-KTEVS SVSSSQVNPS
SWS1_galGa KSACVYNPIIYCFMNKQFR ACIMET-VCGKPLT--DDSDASTSAQRTEVS SVSSSQVGPT
SWS1_xenLa KSSCVYNPIIYSFMNKQFR GCIMET-VCGRPMS--DDSSVSSTSQRTEVS TVSSSQVSPA
SWS1_petMa KASCVYNPLIYSFMNKQFR ARIMET-VCGKFIT--DESETSSS--RTAVS SVSTSQVSPG
SWS1_geoAu KASCVYNPLIYSFMNKQFR ACILET-VCGKPIT--DESETSSS--RTEVS SVSTTQMIPG
SWS1_danRe KSSSVYNPLIYAFMNKQFN ACIMET-VFGKKI---DES--------SEVS SKTETSSVSA
SWS1_oryLa KSSCVYNPLIYAFMNKQFN GCIMEM-VFGKKM---EEA--------SEVS SKTE-VSTDS
P E L
RHO1_hetGl KSSSIYNPVIYIMLNKQFR NCMLTTLCCGKNPLGDDD--ASATASKTETS -----QVAPA
RHO1_musMu KSSSIYNPVIYIMLNKQFR NCMLTTLCCGKNPLGDDD--ASATASKTETS -----QVAPA
RHO1_homSa KSAAIYNPVIYIMMNKQFR NCMLTTICCGKNPLGDDE--ASATVSKTETS -----QVAPA
RHO1_bosTa KTSAVYNPVIYIMMNKQFR NCMVTTLCCGKNPLGDDE--ASTTVSKTETS -----QVAPA
RHO1_monDo KSSSVYNPVIYIMMNKQFR TCMITTLCCGKNPLGDDE--ASATASKTETS -----QVAPA
RHO1_ornAn KSSAIYNPVIYIMMNKQFR NCMLTTICCGKNPLGDDE--ASATASKTEQS SVSTSQVSPA
RHO1_galGa KSSAIYNPVIYIVMNKQFR NCMITTLCCGKNPLGDEDTSAG----KTETS SVSTSQVSPA
RHO1_anoCa KSSAIYNPVIYILMNKQFR NCMIMTLCCGKNPLGDEDTSAGT---KTETS TVSTSQVSPA
RHO1_xenTr KSSAIYNPVIYIVLNKQFR NCLITTLCCGKNPFGDEEGSSAA-SSKTEAS SVSSSQVSPA
RHO1_neoFo KTASVYNPVIYILMNKQFR NCMITTLCCGKNPFGDEETTSA-GTSKTEAS SVSSSQVSPA
RHO1_latCh KSASFYNPVIYILLNKQFR NCMITTLCCGKNPFGDEDATSAAGSSKTEAS SVSSSSVSPA
RHO1_takRu KSAALYNPVIYILLNRQFR NCMITTVCCGKNPFGDDDAATTV--SKTQSS SVSSSQVAPA
RHO1_angAn KSSAIYNPLIYICLNSQFR NCMITTLFCGKNPFQEEE-GASTTASKTEAS SVSS--VSPA
RHO1_conMy KSSALYNPMIYICMNKQFR HCMITTLCCGKNPFEEED-GASATSSKTEAS SVSSSSVSPA
RHO1_calMi KSSALYNPLIYILLNKQFR NCMITTLCCGKNPFEEDE-STSAAASKTEAS SVSSSQVSPA
RHO1_leuEr KSSAVYNPLIYILMNKQFR NCMITTICLGKNPFEEEE-STSASASKTEAS SVSSSQVAPA
RHO1_petMa KTSALYNPIIYILMNKQFR NCMITTLCCGKNPLGDEDSGASTS--KTEVS SVSTSQVSPA
RHO1_letJa KSSALYNPVIYILMNKQFR NCMITTLCCGKNPLGDDESGASTS--KTEVS SVSTSQVSPA
RHO1_geoAu KSSALYNPVIYILMNKQFR NCMITTLCCGKNPLGDDDSGASTS--KTEVS SVSTSQVAPA
P E L
RHO2_galGa KSSSLYNPIIYVLMNKQFR NCMITTICCGKNPFGDEDVSSTVSQSKTEVS SVSSSQVSPA
RHO2_taeGu KSSSLYNPIIYVLMNKQFR NCMITTICCGKNPFGDEETSSTVSQSKTEVS SVSSSQVSPA
RHO2_podSi KSSSLYNPIIYVLMNKQFR NCMITTICCGKNPFGDDDVSSTVSQSKTEVS SISSSQVSPA
RHO2_anoCa KSSSLYNPIIYVLMNKQFR NCMITTICCGKNPFGDEDVSSSVSQSKTEVS SVSSSQVSPA
RHO2_gekGe KSSSIYNPIIYVLLNKQFR NCMVTTICCGKNPFGDEDVSSSVSQSKTEVS SVSSSQVAPA
RHO2_pheMa KSSCIYNPIIYVLLNKQFR NCMVTTICCGKNPFGDEDASSSVSQSKTEVS SVSSSQVAPA
RHO2_neoFo KSSALYNPIIYVLMNKQFR NCMVTTLCCGKNPFGDDDVSSSVSAGKTEVS SVSSSQVSPA
RHO2_latCh KSSCLFNPIIYVLLNKQFR NCMITTLCCGKNPLGDDDTSSAVSQSKTDVS SVSSSQVSPA
RHO2_takRu KSSALYNPVIYVLLNKQFR NCMLSTIGMGGAV--DDE--TSVSASKTEVS -------SVS
RHO2_gasAc KSSALYNPVIYVLLNKQFR NCMLTTIGMGGMV--EDE--TSVSASKTEVS -------SVS
RHO2_oreNi KSSALYNPIIYVLMNKQFR NCMLSTIGMGGMV--EDE--TSVSTSKTEVS -------SVS
RHO2_hipHi KSSALYNPVIYVLLNKQFR NCMLSTIGMGGMV--EDE--SSVSASKTEVS -------SVS
RHO2_mulSu KSSALYNPVIYVMMNKQFR NCILSAIGMGGMV--EDE--TSVSTSKTEVS -------TAS
RHO2_pomMi KSSALYNPVIYVLMNKQFR NCMLSAVGMGGMV--DDE--TSVSASKTEVS -------SVS
RHO2_oryLa KSSALFNPIIYILLNKQFR NCMLATIGMGGMV--EDE--TSVSTSKTEVS -------TAA
RHO2a_danR KTSAVFNPIIYVLLNKQFR SCMLNTLFCGKSPLGDDE-SSSVSTSKTEVS -----SVSPA
RHO2b_danR KASALFNPIIYVLLNKQFR SCMLNTLFCGKSPLGDDE-SSSVSTSKTEVS -----SVSPA
RHO2c_danR KSSSIFNPIIYVLLNKQFR NCMLTTLFCGKNPLGDDE-SSTVSTSKTEVS -----SVSPA
RHO2d_danR KTSALYNPVIYVLLNKQFR NCMLTTLFCGKNPLGDDE-SSTVSTSKTEVS -----SVSPA
RHO2_calMi KSSVLYNPIIYILMNKQFR SSMITTVCCGKNPFGDDD-SSSVTSQSKTEVSSVSTSQVSPA
RHO2_geoAu KSSVLYNPIIYVLLNKQFR TCMVTTLFCGKNPFGEDD-SSMVSTSKTEVS SVSSSQVSPS
P E L
SWS2_ornAn KASTIYNPIIYVFMNKQFR SCMLKLVFCGKSPFGDEDE-ISGSSQATQVS SVSSSQVSPA
SWS2_anoCa KASTVYNPVIYVLMNKQFR SCMLKLIFCGKSPFGDEDD-VSGSSQATQVS SVSSSQVSPA
SWS2_utaSt KASSVYNPVIYVFMNKQFR SCMLKLVFCGKSPFGDEDD-VSGSSQTTQVS SVSSSQVSPA
SWS2_taeGu KASTVYNPIIYVFMNKQFR SCMLKLVFCGRSPFGDEDD-VSGSSQATQVS SVSSSQVSPA
SWS2_neoFo KSSTVYNPLIYVFMNKQFR SCMMKLIFCGKSPFGDEDD-ASSASQSTQVS SVSSSQVAPA
SWS2_galGa KSSTVYNPVIYVLMNKQFR SCMLKLLFCGRSPFGDDED-VSGSSQATQVS SVSSSHVAPA
SWS2_xenTr KASTVYNPFIYIFMNRQFR SCMMKMIFCGKNPLGDDEE--TSVSGSTQVS SVSSSQIAPS
SWS2_takRu KASTVYNPIIYVVLNKQFR SCMKKML---GMSGGDDEE-------SSSQS VTEVSKVSPS
SWS2_gasAc KSSAVYNPVIYVLLNKQFR SCMMKML---GMGGGDDEE-------SSTSS VTEVSKVGPA
SWS2_geoAu KASTVYNPVIYIFLNKQFR SCMMKTIFCGKNPLGDDED---ATSTTTQVS SVSTSQVAPA
P E L
Mole-rat OPN1SW (SWS1) deletional frameshift
The mole-rat assembly exhibits an oddity in exon 4 of STS1 that -- if the sequence is accurate -- is more than sufficient to completely disable this opsin as it takes out the retinal binding site. Note defective opsins not only can still be expressed but also have adverse affects on cone health.
The context however is a string of 6 repeated thymidines tttttt below followed by deletions of 7 and then 3 basepairs relative to the closely related mouse sequence. Since the sequencing technique here involved short reads from an Illumina HiSeq 2000, the assembly may have stumbled in this region.
Below an extra T has been introduced to restore reading frame. This still leaves the retinal lysine followed by a phylogenetically unprecedented 3 bp deletion. The V in 4th position also conflicts with deep invariance. This requires too complex a mutational event or equally implausible multiple events at the same site. This region needs to be resequenced with longer read technology.
ttacggcttgtcaccatccccgccttcttttccaagagctcctgtgtctacaaccccatcatctactgcttcatgaataagcag mouse OPN1SW (SWS1) exon 4 L R L V T I P A F F S K S S C V Y N P I I Y C F M N K Q ttacggcttttttTcctc------ttctcccccaagagc---tgtgtttacaatcccatcatctactgcttcatgaacaagcag mole-rat extra T then 9 bp deletion L R L F F L F S P K S C V Y N P I I Y C F M N K Q
Below, the mole-rat sequence is compared to orthologs from other theran mammals to display anomalies in the deletion region. Otherwise, mole-rat sequences does not seem too unusual. This would require the putative frameshifting deletion to be extremely recent because otherwise the sequence would be much more diverged elsewhere -- and in a manner oblivious to the general sequence conservation pattern. This observation strongly favors sequence error in the TTTTTTT region.
SWS1_hetGla MSGEEDFYLFKNTSSVGPWDGSQYHIAPIWAFHLQAAFMGLVFFVGTPLNAIVLVATLQYKKLRQPLNYILVNVSLGGFLFCIFSVFTVFIASCHGYFFFGRHVCALEAFLGSVAGLVTGWSLAFLAFERYLVICKPFGNFRFSSKHALIVVLATWVIGIGVSIPPFFGWSR SWS1_musMus ....D.....Q.I........P...L..V...R.......F.................H.......................................L................................V.......SI..N.....M......I............... SWS1_ratNor ....DE....Q.I........P......V...........F...A......T......H.......................................L.........................................I..N.....T...I..T............... SWS1_criGri ....DE......I........P......V...........F..LA......T...V..R.......................S...............L................................I........I........M...V..T............... SWS1_cavPor ..E.........A........P...V..V...R.......I..CI.....G.......L.....................V..................L................M...................................................... SWS1_homSap MRKMS..E......I........P......V...Y.......T..LI.F....M......R................F....L......P..V...N...V.........G...T..................I.................T......T............... SWS1_oryCun RNTMS..E......L.T.R....P.....R-...........F..........M......R..............I..A...A......N..V...Y...V...F.......T..A.............L...II................L...V..T..V............ SWS1_ochPri RSTMS..E......L...A....P......................A......V......R..............I..A...A......N..LS..Y...VL..F..S....T..AT................I.................V.........V...M........ SWS1_panTro MRKMS..E......I........P......V...Y.......T..LI.F....M......R................F....L......P..V...N...V.........G...T..................I.................T......T............... SWS1_gorGor MRKMS..E......I.P......P......V...Y.......T..LI.F....M......R.......X........F....L......P..V...N...V.........G...T..................I.................T......T............... SWS1_ponAbe MRKMSD.E......I........P......V...Y.......T..L..L....M......R................F....L......P..V...N...V.........G...T..................I.................T......T............... SWS1_rheMac MRKMS..E......I...K....P......V...Y.......T..LA.F....M.....VR................F....L......P..VN..K...V.......F.....T..................I.................T......T............... SWS1_papHam MRKMS..E.F....I...K....P......V...Y.......T..LA.F....M.....VR................F....L......P..VN..K...V.......F.....T..................I.................T......T............... SWS1_calJac MSKMS..E......I........P......S..YY.......I..LI.L...TM.....VR.....H....V.....V....L......P..V.......V.........G...T..................I.................M...T..T............... SWS1_tarSyr MSKMS..E......I........P......S.V..I......V..S..I....M......H.R...................L.....LP......R...V.........G......................M............P..S.M......T......V........ SWS1_micMur MSKMS..E......L........P......V.T.Y...V...F...A.....VM......R..............L.F....S.....LP......R...L.........G...CA....I............V.............R...M......A............... SWS1_otoGar MSKMS..E......L........P...L..V...C.E.-...F...A.....VM......C....W...S....SL....I.N..L..LPI.....-...L..C.A....G..S..V...I.-........C.I.........C....P..M......TT...I..ST...... SWS1_tupBel ISKMS..E......A.L......P...L..V...........F..........S.....MR.R............I.F....Y......V..LN..........FI.G....M.T.....I.-........C.I.........C....P..M......TT...I..ST...... SWS1_vicPac ..KMS..E......I...K....P...L......R.......F..L.......T..I...R.R..................IY.M....C..V...Y...V...R...M......A.................II...............VM..V...I..V............ SWS1_turTru MSKMS..E.-....I--......P..RL..V.........T.F..V.....D.T......R.R..................IY......V...T......V........G....RT...L........V....II................M......T............... SWS1_bosTau MSKMS..E.L....I.L......P...L..V.......V...F..........T......R.R..................IY......I...T..Y...V.............CT.................II................M..V...T............... SWS1_equCab MSKMS..E.F.Y..I...R....P......V...R.....L.I..L..M...SL......R.....................V.....LI...N......V..P....F.S...T..................II................M......I...S........... SWS1_felCat MSKMSG.E......I.L......P......V.......V...F..........T......R.R...................Y.VS..SI...T...A..I.......W...M.CT...................................M......T............... SWS1_canFam MSKMSG.E......I.L......P......V.......V...F...A.....GT......R.....................Y..V.--.......Q...V..............T.................I.................M......T............... SWS1_myoLuc MSKMSG.E......I........P......V...........F...A......T......R...................................Q...V..............T.................I.V.....S................T............... SWS1_pteVam MSKMSG.E......I.L......P.H....V...........F..........T......R.R.................................Q...V..............T.................I.................M......T............... SWS1_eriEur INKMSG.E......I........P......L...........F...A.....SM..L...R.R.........................L.......K...I..PL...W......A.....S...........V.V.....S.....R...M......IM.V............ SWS1_sorAra .SKMSG.E......I.T......P......V...........F...A......T...P..R.R............................I....K...VI........T..A.A.................II................L...................... SWS1_loxAfr MSKMSG.E......I......G.P....G.V..........AF..........L......R.R............I......S......I...S..K...I...Y.......V....................I........I........M......T............... SWS1_monDom .....D.E......I........P......A....F.TV...F..CA......V......R.................C..I....A......S.SQ...I.......M.......................FI...........N....MM...................... SWS1_smiCra .....D.E......I.L......P...L..A....F.T....F...A..S..GV..I...R..............I..A..I..V.......VS.SQ...V.......M.G.....................FI...........N....MM......I............... SWS1_tarRos .....D.E.....DI........P......A....F.TT...F...A......V..I...R..............I..A..I..VI.......S.SQ...I.......M.......................FI................MM...................... SWS1_hetGla YIPEGLQCSCGPDWYTVGTKYRSEYYSWFLFIFCFIVPLSLICFSYSQLLRTLRAVAAQQQESATTQKAEREVSRMVVVMVGSFCVCYVPYAALAMYIVNNRNHGLDLRLFFL--FSPKS-CVYNPIIYCFMNKQFRACIMELVCRKSMADESDMSSSQKTEVSAVSSSKVGPN SWS1_musMus F.........................T.........I.....................................H..........L...........M............V.......KS....................L.M....P......V.G.......T........H SWS1_ratNor F.......................H.T.........I.........F...........................H..........L...........M............V.......KS....................L.M....P.T......G.......T........H SWS1_criGri F.........................T....L..........................................H..........L...........M............V.......KS......................M....PVT......G.....A.T........H SWS1_cavPor .M..............M........FA...................C.......T.......................................................V.......KS..I........................P........T................. SWS1_homSap F.......................S.T...................T....A.K.......................................F...M............V...S...KSA.I.............Q....KM..G.A.T....TC........T...TQ.... SWS1_oryCun F.........................T.........I...I.....A.............................................S....M............V.......KS.................G....M..G.P.T.D..V..A......T....R.... SWS1_ochPri FL........................T.........I.........A...G.........................................S................FV.......KSA................G....M..G.P.T.D..V..A......T....R.... SWS1_panTro F.......................S.T...................T....A.K.......................................F...M............V...S...KSA.I.............Q....KM..G.A.T....TC........T...TQ.... SWS1_gorGor F.......................S.T...................T....A.K.......................................F...M............V...S...KSA.I............VGPN..KM..G.A.T....TC........T...TQ.... SWS1_ponAbe F.......................S.T...................T....A.K.......................................F...M............V...S...KSA.I.............Q....KM..G.A.T....TC........T...TQ.... SWS1_rheMac F.......................S.T...................T....A.K.......................................F...M............V.......KSA.I.............Q.H..KM..G.A.T....I.........T....Q.... SWS1_papHam F.......................S.T...................T....A.K.......................................F...M............V.......KSA.I.............Q.H..KM..G.A.T....I.........T....Q.... SWS1_calJac ..A.......................T............A......A....A.K...........................................M............V.......KS..I......S............M..G.A.T....I.........T....Q.... SWS1_tarSyr F.........................T.........M.........A....A......................C..........L...........M............V...S...KSA.I.............Q.....M....A......T..........L...Q.S.. SWS1_micMur F................D........T....L..................QA.................................L........................V.......KSA...............Q.....M..G.A.T....T.......I.TF...Q...K SWS1_otoGar F...........NY.-M....YH...T..........LI......S....GAR.D...........H.......H........S.--..........M...C......L.V.......KSA...............Q.....M....ALT...NIF.P......IL...H...I SWS1_tupBel F.........................T............A..........GA........................LM..-................M.......I....V.......KSA.....................M....P.T...E.....R....T....Q.... SWS1_vicPac .L...................Y....T.......Y...............GA............S.........H...M......L..T................V....V.......KSA.....................M..G...T..............T....Q.... SWS1_turTru FA.......------......Y....T.......YT..............GVF.........A......G....HK..M......L..T............H...V...FV..S...PKSA...............Q.....M..G-P.T..........M...T....Q.... SWS1_bosTau FV...................Y....T.......Y...............GA............S.........H..........L..T................V....V.......KSA.....................M..G.P.T...EL.........T....Q.... SWS1_equCab F.................................................GA.............................................M...P.......FV.......KS......................M..G.A.I...EL.I.......T....Q.... SWS1_felCat F.........................T...................M...GA..V-------..S.......................T....M...M............V.......KSA.....................M..G...T.D.EV..........L...Q.... SWS1_canFam F.........................T...................L....A............S....................L..T....M...M............V.......KSA.....................M..G...TED.E..........T..P.Q.... SWS1_myoLuc F.........................T.......................GA............S....................L...........M............V.......KSA..................M..M..G...T..............TL...Q...S SWS1_pteVam F.........................T.......................GA............S....................L...........M............V.......KSA...............Q........G.P.T.........R....TL...Q.... SWS1_eriEur F....................H..S.T....L....A.........L..........V......S.............M......L..........F.....D.......V.......KSA........R............M..G...I.D........M...S..P.-ID.. SWS1_sorAra F.........................T.......................GA............S....................L...........M....D.......V.......KSA.....................ML.GR.VTED..TP........TL...Q.... SWS1_loxAfr FL........................T.......................GA.............................A...L...........M............V.......KSA................G....M..G..V...........M...T....Q.... SWS1_monDom F.........................T.........M..F...........A..........................M......L...........M...Q........V.......KSA...............H.....M....P.T.D..V..............Q...T SWS1_smiCra ..........................T.......................GA.................................L.......M...M............V.......KSA...............H.....MI.K.P.T.D.ETT........T....Q...S SWS1_tarRos .....................H....TG......................GA.................................L...........M............V.......KSA......V.W......H.....M....P.T.D.EI.........T....Q...S
Alignment of mole-rat to 4 other subterranean hystricognath rodents and lawn mole: SWS1_hetGla LQAAFMGLVFFVGTPLNAIVLVATLQYKKLRQPLNYILVNVSLGGFLFCIFSVFTVFIASCHGYFFFGRHVCALEAFLGSVAGLVTGWSLAFLAFERYLVICKPFGNFRFSSKHALIVVLATWVIGIGVSIPPFFGWSRYIPEGLQCSCGP SWS1_cteMag ...S...F...A.............K.......................V...S...........L...E..............................................M.................................. SWS1_cteTal ...S...F...A.............K.......................V...S...........L...E............--------------------------------------------------------------------- SWS1_spaCyn .......F...A.....................................................L..HQ........................L.....................M.................................. SWS1_octDeg .......F...A.....................................................L..HQ........................L.....................M.............................----- SWS1_cavPor .......I..CI.....G.......L.....................V.................L..HQ.............M........................................................M.......... SWS1_talEur ----------.A......T......R.R.........................L.......K...I..............A.................I..........------------------------------------------ cteTal Ctenomys talarum (tuco-tuco) ADF36522 Hystricognathi spaCyn Spalacopus cyanus (coruro) ADF36520 Hystricognathi octDeg Octodon degus (degu) ADF36519 Hystricognathi [lives in burrows but good vision] cavPor Cavia porcellus (guinea_pig) Hystricognathi [not subterranean but hystricognath] cteMag Ctenomys magellanicus (tuco-tuco) ADF36521 Hystricognathi talEur Talpa europaea (mole) ABV31135 Laurasitheres Insectivora
Cryptochromes CRY1 and CRY2 in mole-rat
The primary evolutionary adaptation to a rotating planet and its latitudinal gradients is accomplished by cryptochrome genes. In mammals, CRY1 and CRY2 mediate circadian rhythm entrainment with many downstream effects on metabolism and additionally magnetic field orientation though they have lost former photolyase capacity to repair UV damage to dna (thymine dimers and 6-4 fusions).
Their continuing importance to a subterranean rodent -- previously studied in a murid blind mole-rat Spalax -- is implied by degree of continuing sequence conservation relative to genes being lost. Although daily rhythms remain relevant to metabolism and so longevity even when imaging vision is not, magnetic field orientation could conceivably be applicable to dark tunnel navigation.
CRY 1 is localized to mitochondria via a targeting sequence, whereas its paralog CRY2 is expressed, among other places, in inner retina where the putative FAD radical sensor could overlay a magnetic field direction on the opsin output. Note melanopsin is often ascribed the primary circadian rhythm photoreceptor function but could interact synergistically with CRY2.
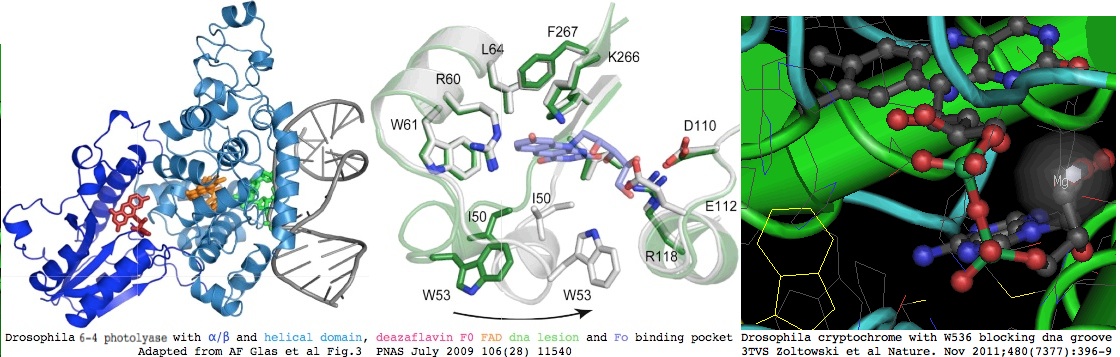
The extreme conservation of CRY2 would enhance the relevance of extensive studies in the Drosophila model system but functional correspondences drifted off after separate lineage-specific duplications. Drosophila encodes three paralogous cryptochromes and photolyases: a cyclobutane pyrimidine dimer photolyase (called CPD_droMel here), a pyrimidine-pyrimidone 6-4 photolyase (CRY64_droMel), and a CRY1B-type cryptochrome. The first two use 5-deazariboflavin as their second chromophore but none of the known antenna molecules prove applicable to the third. The key enzyme that synthesizes deazariboflavin persist into land plants (mosses and club-mosses; Physcomitrella patens XM_00177156, Selaginella moellendorffii XM_002968187) but otherwise this molecule is a vitamin in species such as Drosophila that have lost the enzyme. Deuterostomes may use alternative antenna molecules or nothing at all.
The structural fold has been determined for both CRY64_droMel (notably 3CVW) and very recently for CRY1_droMel (3TVS), leaving vertebrate CRY2 without a direct representative. The latter has a C-terminal alpha helix docked in what is normally an open groove that binds the dna lesion in photolyases, with W536 serving as lesion proxy in the catalytic center and the anionic semiquinone form of FAD facilitating this rearrangement. Thus the fold, including the dna binding groove, is conserved but cryptochromes are not catalytically active in dna repair nor capable of site-specific dna recognition despite their role in regulation of gene expression. There is no counterpart to W536 in vertebrate CRY1 or CRY2.
Mole-rat CRY2 is 53% identical to Drosophila CRY64, 39% identical to CRY1B but greatly diverged from CPD -- thus the fold of CYR64 is a better for structural modeling of mammalian CRY2 proteins than Drosophila CRY1B. The mole-rat difference alignment of CRY2 is shown below for placental mammals. Apart from a few compositionally simple regions with rapidly evolving underlying DNA, CRY2 is extraordinarily conserved with only a few exceptional sites in mole-rat.
Here the photolyase domain is shown in brown, the FAD binding region blue, the CLOCK-ARNTL transcription inhibition region in green, the tryptophan triad supposedly central to electron transport but now disputed in red and key residues of F0 (and presumably MTHF) in magenta.
Note the Pfam approach of breaking down proteins into linear contiguous domains does not so work well for this protein -- disjunct peptides fold back to form to individual functional domains and other structural units are scattered across multiple domains. The regions are highly conserved in mole-rat CRY2, indeed so highly that triad tryptophans do not stand out among other residues for an exceptional degree of conservation (which is largely unexplained to date).
The difference alignment of mammalian CRY1 sequences also exhibits extraordinary conservation except for the last two exons in hystricognath rodents. Mole-rat itself is a pseudogene as the assembly stands because a stop codon occurs a position 8 (in place of an invariant tryptophan) and no later methionine is suitable for initiation. The change, TGG to TAG, is not at a CpG hotspot. Sequence error could be the problem here because the rest of the protein seems unaffected (other than the odd divergence C-terminally -- but that is also seen in guinea pig).
However the independent sequencing of a different (female) individual by Broad shows this same stop codon at position 8 of CRY1. See AHKG01086374 positions 2900-3079. Thus it cannot plausibly represent a sequencing error or mutation in the BGI individual but instead most likely occurs homozygously across a population. Loss of CRY1 is unprecedented in placental mammals (though mouse knockouts are known and not lethal).
The mole-rat genome also contains several processed CRY1 pseudogenes. The longest of these (AHKG01030319) does contain a tryptophan at position 8: ...HWFRKGLWLHNNPALKECIRGADTIHSVHIL. This not only implies significant historic transcription in germ line cells, presumably testis, as well as fairly recent acquisition of the stop codon. This fits the notion of CRY1 loss being a recent beneficial adaptation to the conditions of underground metabolism.
Lab mouse has an odd mutation in its 10th exon where a century of inbreeding may have inadvertently fixed a very serious 54 bp tandem stutter mutation resulting in 18 additional amino acids (NGGLMGYAPGENVPSCSGG in NM_007771 reference sequence) that would very likely disrupt the protein fold (however this distal region is not modeled in any PDB structure as of April 2012). A mutation in this critical pacemaker gene could plausibly affect lifespan, metabolic disorder and tumor progression; such a change is completely unprecedented in rodents including rat and indeed in vertebrates. The repeat was first noticed in 1998 but could not be recognized as a mutation because comparative genomics then was non-existent.
All 14 available transcripts exhibit the same anomaly -- this is limited to one strain of mouse, nor a somatic mutation, nor an unfortunate heterozygous allele. The affected ESTs came from C57BL/6J, C57BL/6, C57BL/6J x DBA/2J, 129 FVB/N and embryo, eye, ventricle, thymus, mammary tumor; the affected GenBank NR entries add a keratinocyte cell line Pam. The mouse genome project used C57BL/6J, the most widely used inbred strain according to the Jackson Laboratory:
"Although C57BL/6J is refractory to many tumors, it is a permissive background for maximal expression of most mutations. C57BL/6J mice are resistant to audiogenic seizures, have a relatively low bone density, and develop age related hearing loss. They are also susceptible to diet-induced obesity, type 2 diabetes, and atherosclerosis. C57BL/6J mice are used in a wide variety of research areas including cardiovascular biology, developmental biology, diabetes and obesity, genetics, immunology, neurobiology, and sensorineural research. C57BL/6J mice are also commonly used in the production of transgenic mice. Overall, C57BL/6 mice breed well, are long-lived, and have a low susceptibility to tumors. Primitive hematopoietic stem cells from C57BL/6J mice show greatly delayed senescence relative to BALB/c and DBA/2J. This is a dominant trait. Other characteristics include: 1) a high susceptibility to diet-induced obesity, type 2 diabetes, and atherosclerosis; 2) a high incidence of microphthalmia and other associated eye abnormalities; 3) resistance to audiogenic seizures; 4) low bone density; 5) hereditary hydrocephalus (early reports indicate 1 - 4 %); 6) hairloss associated with overgrooming, 7) a preference for alcohol and morphine; 8) late-onset hearing loss; and 9) increased incidence of hydrocephalus and malocclusion."
2 GLLASVPSNSNGNGGLMGYAPGENVPSCSSSGNGGLMGYAPGENVPSCSGG 1 exon 10 in mouse CRY1
G N G G L M G Y A P G E N V P S C S
gggaatggagggctcatgggctatgctcctggagagaacgtcccgagctgtagc first repeat
gggaatggagggctcatgggctatgctcctggagagaatgtcccgagttgtagc second repeat
G N G G L M G Y A P G E N V P S C S
507 517 527 537 547 557 567 577 587 597
| | | | | | | | | |
CRY1_musMus GLLASVPSNSNGNGGLMGYAPGENVPSCSSSGNGGLMGYAPGENVPSCSGG NCSQGSGILHYAHGDSQQTHSLKQ GRSSAGTGLSSGKRPSQEEDAQSVGPKVQRQSSN*
CRY1_ratNor GLLASVPSNPNGNGGLMGYAPGENVPSGGSGG------------------G NCSQGSGILHYAHGDSQQTNPLKQ GRSSMGTGLSSGKRPSQEEDAQSVGPKVQRQSSN*
CRY1_criGri GLLASVPSNPNGNGGLMGYTTGENLPSCSGGG------------------- SCSQGSGILHYAHGDSQQAHLLKQ GRSSMGTSLSSGKRPSQEEETRSVDPKVQRQSSN*
CRY1_spaJud GLLASVPSNPNGNGGLMGYTPGENIPNCSSSG------------------- SCSQGSGILHYAHGDSQQAHLLKQ GSSSMGHGLSNGKRPSQEEDTQSIGPKVQRQSTN*
CRY1_dipOrd GLLASVPSNPNGNGGLMGYAAGDNLPGSSSSG------------------- SCSQGSGILHYAHGDSQQMHLLKQ GRSSMGTGLSSGKRPSQEEDSQSIGPKVQRQSTN*
CRY1_hetGla GLLASVPSNPNGNGGLMGYAPGESIPGSSGSG------------------- SCAHGSGILPCAHTDGQQAHLLKP GRNCVGPVLSSGKRPSQEEDAQSIGPKLQRQSTD*
CRY1_cavPor GLLASVPSNPNGNGGLLGYAPGESTPGSGGG-------------------- SCVPGSSSAGVSHCAQGEAPQAPP GRDPAGPGLGGGKRPSQEEDAQSTGHKIQRQSPD*
CRY1_speTri GLLASVPSNPNGNGGLMAYAPGENIPGCSSSG------------------- SCTQGSSILHNAHGDSQQTHLLKQ GRSSMGTGLSSGKRPSQEEDTQSIGPKVQRQSTN*
CRY1_oryCun GLLASVPSNPNGNGGLMGYSPGENIPGCSSSG------------------- SCSQGSGILHYAQGDTQQTQLLKQ GRSSMGTGLSSGKRPSQEEDTQSIGPKVQRQSTN*
CRY1_musMus GLLASVPSNSNGNGGLMGYAPGENVPSCSSSGNGGLMGYAPGENVPSCSGG NCSQGSGILHYAHGDSQQTHSLKQ GRSSAGTGLSSGKRPSQEEDAQSVGPKVQRQSSN*
CRY1_ratNor .........P.................GG.G.------------------. ...................NP... ....M.............................
CRY1_criGri .........P.........TT...L....GG.------------------- S.................A.L... ....M..S...........ETR..D.........
CRY1_spaJud .........P.........T....I.N.....------------------- S.................A.L... .S..M.H...N.........T..I........T.
CRY1_dipOrd .........P..........A.D.L.GS....------------------- S.................M.L.... ...M...............S..I........T.
CRY1_hetGla .........P.............SI.GS.G..------------------- S.AH.....PC..T.G..A.L..P. .NCV.PV...............I...L....TD
CRY1_cavPor .........P......L......ST.GSGGG-------------------- S.VP..SSAGVS.CAQGEAPQAPP. .DP..P..GG............T.H.I....PD
CRY1_speTri .........P.......A......I.G.....------------------- S.T...S...N.........L.... ...M...............T..I........T.
CRY1_oryCun .........P.........S....I.G.....------------------- S...........Q..T...QL.... ...M...............T..I........T.
CRY2_hetgla MAAAVGTGTGAAPTPATGAEGACSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRWRFLLQSLEDLDTSLRKLNSRLFVVRGQPADVFPRLFKEWGVTRLTFEYDSEPFGKERDAAIMKMAKEAGVEVVTENSHTLYDLDRIIELNGQKPPLTYKRFQAIISRMELPKKPVGAVSSQQMKSCR CRY2_cavPor .........A.....V..................................................................................................................................................................................E... CRY2_musMus ....AVVA ATV.AQSM..D..S.......................................................................................................................................................L..........AV.......E... CRY2_ratNor ....AVVA ATV.AQSM..D..S.......................................................................................................................................................L.................H.EN.. CRY2_dipOrd ....MV.AAV.V.A.PS..D..S....................................................................................................................................................................S.T....E... CRY2_speTri .S.S.V.TSATLL..TSA DVS........... ................................................................................................................... ......................................E..K CRY2_oryCun ....AAAAAA.V.A..AS.N..S............................................................................T.....................................................................................A.S.T...LEG.. CRY2_ochPri ...TAAAA V..AA. ..S................................................ ............................R.............................................R....S.T.E..E... CRY2_homSap ...T.A.AAAV..A..P.TDS.S....................................................................................................................................................................L.T....E... CRY2_panTro ...T.A.AAAV..A..P.TD..S....................................................................................................................................................................S.T....E... CRY2_gorGor ...T.A.AAAV..A..P.TD..S..................................................................................... ...............................S.T....E... CRY2_ponAbe ...T.A.AAAV..A..P.TD..S....................................................................................................................................................................S.T....E... CRY2_rheMac ...T.A.AAAV..A..P.TD..S...................................................................................................................................E................................S.T....E... CRY2_papHam ...T.A.AAAV..A..P.TD..S...................................................................................................................................E................................S.T....E... CRY2_calJac ...T.A.AAA.V.A..P.TD..S....................................................................................................................................................................S.T....E... CRY2_otoGar .....A.A .QG...S.D..S............................................................................................................................ ...............................S.T....E... CRY2_vicPac ..T..A.AAA...A..A.....T........................H...........................................................................................................................................S.T....E... CRY2_turTru .....A.SAV...A..AR....S.................Q......H...........................................................................................................K...............................S.T....EG.. CRY2_bosTau ....AAA TQA..ARGD..S........................H....................... .....................T.........................K..........................R....S.T....EG.. CRY2_equCab ...............................................................................................................R....S.T....E... CRY2_felCat ...A..A..D..S.......................................................................................................................................D............................S.T....E... CRY2_canFam .....VAAAA...V.TA.VD..S....................................................................................................................................K...............................S.T....E... CRY2_myoLuc ...NAV.AAA...A..A.TD..S..Y........................L.......................................................................................................................................AS.TRH..E..P CRY2_pteVam ...T...AAA..SA..A.TD..S............................................................................T.......................................................................................S.T..H.E..T CRY2_eriEur ...VAV.AAA..AG.GPV.D..S.........................S...................... .....................T....................................................R....S.T....E... CRY2_sorAra ..T.AAAAAA...A..AR.......................................................................................... ....................G.....M....S..R...E... CRY2_loxAfr .....V.AGA..LV.IPSMD..S....................................................................................................................................................................S.T....E... CRY2_proCap ...TAA.AVA..LA..PT.D.VS......................................................................................A. R.................................V......R....S.T....E... CRY2_echTel .......AAL..TA..PNKD..S.......................................................A...AR..T.H.............................................L...............................................R..A..PTGL..E..Q CRY2_hetgla AEIQENHDDTYGVPSLEELGFPTEGLGPAVWQGGETEALARLDKHLERKAWVANYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLWDLYKKVKRNSTPPLSLFGQLLWREFFYTAATNNPRFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWESG CRY2_cavPor ...................................................................................................................................................................................................... CRY2_musMus ...................................................................................................................................................................................................... CRY2_ratNor ..............................................................................................R....................................................................................................... CRY2_dipOrd ........................................................................................................................................................................ .............T............ CRY2_speTri .....D............................................................................................K................................................................................................... CRY2_oryCun ....D...E.................S........................................................................................................................................................................... CRY2_ochPri ....D...ES......... .......................................K..S.....Y....................K.......V.....K...........R.....................................I...E. CRY2_homSap ........E............................................................................................................................................................................................. CRY2_panTro ........E............................................................................................................................................................................................. CRY2_gorGor ........E............................................................................................................................................................................................. CRY2_ponAbe ........E............................................................................................................................................................................................. CRY2_rheMac ........E............................................................................................................................................................................................. CRY2_papHam ........E............................................................................................................................................................................................. CRY2_calJac ........E............................................................................................................................................................................................. CRY2_otoGar ........E.......... ..................................................................................................................................................... CRY2_vicPac ........E.F........ ...................................................................................................... CRY2_turTru ........E..........................................................................................S.................................................................................................. CRY2_bosTau .....S..E............................................................................................................................................................................................. CRY2_equCab .D......E.....................................................A...............................R..........................................................A............................................ CRY2_felCat ........EA...................................... ....................................................................................................... CRY2_canFam .D..D................................................................................................................................................................................................. CRY2_myoLuc ........E............................................................................................S................................................................................................ CRY2_pteVam ........E............................................................................................................................................................................................. CRY2_eriEur ........E............................................................................................................................................................................................. CRY2_sorAra ........E........G.............................. ....SH........K.P...L ..S...H..S.... .....PD........................................................... CRY2_loxAfr ........E..................................................................................E.......................................................................................................... CRY2_proCap ..M....EEI...................................... ....................................................................................................... CRY2_echTel ...P...AE............................................S....................................................................T........................................................................... CRY2_hetgla VRVFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYIRRYLPKLKGFPSRYIYEPWNAPESVQKAAKCIIGVDYPRPIVNHAETSRLNIERMKQIYQQLSRYRGLCLLASVPSCVEDLSHPVAEPTLSQAGSSSSTGPRSLPDGPASPKRKLEAAEEPPGEELSKRARVAELPAPEPTSKDA CRY2_cavPor ............................................................................I...............................................................S.......T.....P..G..............................T...P.... CRY2_musMus ............................................................................................................................................GS.....I.N....A.SS....................T.....T.M.TQ..A...S CRY2_ratNor ............................................................................................................................................GS.....I.N....P.SS..........................T.M..Q..P...S CRY2_dipOrd ....R..................I...............................................................SS.....I......P..SS................................LP...S CRY2_speTri ............................................................................I...............................................................GS.....I......P.SS.Q............................T..LP.... CRY2_oryCun ...................................................V........................................................................................SS.....V.GAA..P..S............................V.T..LP..AV CRY2_ochPri MK ...........................................................................I.....................................................SS....NV......P.SS...........................VV....LP..AV CRY2_homSap ............................................................A...............I...............................................................SS.....M..A...P..S..............................T..LP.... CRY2_panTro ............................................................A...............I...............................................................SS.....M..A...P..S..............................T..LP.... CRY2_gorGor ............................................................A...............I...............................................................SS.....V..A...P..S..............................T..LP.... CRY2_ponAbe ............................................................A...............I...............................................................SS.....V..A...P..S..............................T..LP.... CRY2_rheMac ............................................................................I...............................................................SS.....VN.A...P..S........................K.....T..LP.... CRY2_papHam ............................................................................I...............................................................SS.....VN.A...P..S........................K.....T..LP.... CRY2_calJac ............................................................................I...............................................................SS.....M..A...P..S..............................T..LP...V CRY2_otoGar ................S...........................................................I....................................................M..........CS.....INN....P.LS.S........................T...T..LP.... CRY2_vicPac ............................................................................................M.........................................N.....SS.....M..A...P..G..............................ST.LPGR.V CRY2_turTru ............................................................................I....................................................M....N.....SS....GM..A...P..S.........G.................G..PS.LL...V CRY2_bosTau ............................................................................I......V..............A..........V........................N.....SSI.......V...P..G.................G..........SLPS.LP.RGV CRY2_equCab ............................................................A..............A..........................................................N.....SS.....V..A...PP.S.......................... .P.SRAV . CRY2_felCat .........................................................M..................I.........................................................N.....SS..T.NV..A CRY2_canFam .........................................................I..................I.........................................................N.....SS..T..M..A...P..S..............................T..LPCR.V CRY2_myoLuc F...........................................................................I....................................................M....N.....S...T..M..A..KP..S...............................T.LP.R.V CRY2_pteVam ............................................................................I........................................................NN.....SS..T..MNNA...P..S.................................LP.R.V CRY2_eriEur ......................................................................................................................................N.M...SS.....A..A.....TS........PKT..... CRY2_sorAra ...........................................................................A.......V........M......N..T.........K..S.W.H...........E..N.M...SG......A CRY2_loxAfr ............................................................................I.........................................................S.....SS......N.A...P..S..T........................K..G..LP...V CRY2_proCap ............................................................................I.........................................................N.....SS.P..HM..A.L.P..S...........................K..G..LP.T.. CRY2_echTel ....................................................
CRY1_hetGla MAMNAVH*FRKGLRLHDNPALKECTRGADTIRCVYILDPWFAGSSNVGINRWRFLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFKEWNITKLSIEYDSEPFGKERDAAIKKLATEAGVEVIVRISHTLYDLDKIIELNGGQPPLTYKRFQTLISKMEPLEIPVETITSEVIEKCVTPLSDDHDEKYGVPSLEELGFDTDGLPSAVWPGGETEALTRLERHLERKAWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKKVKKNSSPPLSLYGQLLWRE CRY1_musMus .GV....W................IQ..................................................................................................................................V.......M.AD....D..G..M..........................S........................................................................................ CRY1_ratNor .GV....W................IQ..................................................................................................................................V.......M.AD....D..G..T..........................S........................................................................................ CRY1_dipOrd .GV....W................IQ........................................................................................................................................................T..........................S........................................................................................ CRY1_cavPor .GV....W................I............................................................................................S........................................................V....................................................................................................................... CRY1_speTri . .................R ......................................................................T..........................S........................................................................................ CRY1_oryCun .GV....W................IQ....................................................................R......................S......................................................D.....T..V.......................S........................................................................................ CRY1_homsap .GV....W................IQ........................................................................................................................................................T..........................S........................................................................................ CRY1_panTro .GV....W................IQ........................................................................................................................................................T..........................S........................................................................................ CRY1_ponAbe .GV....W................IQ......................................S.................................................................................................................T..........................S........................................................................................ CRY1_rheMac .GV....W................IQ........................................................................................................................................................T..........................S........................................................................................ CRY1_papHam .GV....W................IQ........................................................................................................................................................T..........................S........................................................................................ CRY1_calJac .GV....W................IQ........................................................................................................................................................T..........................S........................................................................................ CRY1_otoGar .GV....W................IQ....................................................................R..............................................................................I....T...F............... .................................................................. CRY1_vicPac .GV....W................IQ........................................................................................................................................................T................................................................................................................... CRY1_turTru .GV....W.............. .IQ........................................................................................................................................................T..........................S......................KF .... ........ CRY1_bosTau .GV....W................IQ........................................................................................................................................................T................................................................................................................... CRY1_equCab .GV....W................IQ........................................................................................................................................................T.....................................................................A..................R.....R.................... CRY1_canFam .GV....W................IQ....................................................................................................................................................V...T................................................................................................................... CRY1_myoLuc ..W.............L..AGR. V...........A..S.................S..............................................................Q............................................L......T................................................................................................................... CRY1_pteVam .GV....W................IQ...................................................................A....................................................................................T................................................................................................................... CRY1_sorAra .GV...PG....AA...K...C..IQ...S.G....S..RY....E ..TK..................................................................................... ............................................................................................... CRY1_loxAfr .GV....W................IQ.................................................................D...................................................................................G..T................................................................................................................... CRY1_proCap .GV....W................IQ..............................R....... .... .Q...... ............D.................................................................................L....T...C...................................................... .. ....................... .......... CRY1_echTel .GV....W................IQ.............................................................. .............................................................................................................................................................. CRY1_dasNov .GV....W................IQ.......................... ..................................................................................P.M....T................................................................................................................... CRY1_choHof .GV....W................IQ.....................................SS...S..H...TH ...... ...HV..........T..............Q....FX... ........L........................IP.L....T................V.. ..........X.........H........ ........Q..........I..M.....................I..............H..... CRY1_hetGla FFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWEEGMKVFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIRRYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGLGLLASVPSNPNGNGGLMGYAPGESIPGSSGSGGCAHGSGILPCAHTDGQQAHLLKPGRNCVGPVLSSGKRPSQEEDAQSIGPKLQRQSTD- CRY1_musMus .............................................................................I......................................................................................................................................S.......... ..NV.SC.SG.N.SQ.....HY..G.S..T.S..Q..SSA.TG...............V...V....SNW CRY1_ratNor .............................................................................I................................................................................................................................................. ..NV.SGGSG.N.SQ.....HY..G.S..TNP..Q..SSM.TG...............V...V....SNW CRY1_dipOrd .............................................................................I..........................................S...................................................................................................... .DNL....S..S.SQ.....HY..G.S..M....Q..SSM.TG............S......V.....NX CRY1_cavPor ...........................R.....................................................................................................................................................H.........................................L... ...T.. ..G.S.VP..S.SH..QGEAP.. ...DPA..G.GG............T.H.I....P.. CRY1_speTri .............................................................................I..................................................................................................... ... .............. ............AX. ..N...C.S..S.TQ..S..HN..G.S..T....Q..SSM.TG............T......V.....NX CRY1_oryCun .......................................Q.....................................I...........................................................................................I....................................................S ..N...C.S..S.SQ.....HY.QG.T..TQ...Q..SSM.TG............T......V.....NX CRY1_homsap .............................................................................I............................................................................................................................................F...S A.N...C.S..S.SQ.....HY..G.S..T....Q..SSM.TG..G.........T......V.....NX CRY1_panTro .............................................................................I............................................................................................................................................F...S A.N...C.S..S.SQ.....HY..G.S..T....Q..SS..TG..G.........T......V.....NX CRY1_ponAbe .............................................................................I............................................................................................................................................F...S A.NV..C.S..S.SQ.....HY..G.S..T....Q..SSM.TG..G...A.....T......V.....NX CRY1_rheMac .............................................................................I...........................................................................................I................................................F...S T.N...C.S..S.SQ.....HYT.G.S..T....Q..SSM.TG..G.........T......V.....NX CRY1_papHam .............................................................................I...........................................................................................I................................................F...S T.N...C.S..S.SQ.....HYT.G.S..T....Q..SSM.TG..G.........T......V.....NX CRY1_calJa1 .............................................................................I............................................................................................................................................F...S A.N...CTS..S.SQ.....H...G.S..T....Q..SSMSTGI.G.........T......V.....NX CRY1_otoGar .............................................................................I...........................................AS..............................................I...............................................SF.E.S P.N...C.S.. . CRY1_vicPac .............................................................................I...........................................................................................................................................S....S ..N...C.S..S.TQ..S..HY..G.S.......Q.KSST.TG............M....S.V.....NX CRY1_turTru .............................................................................I................................................................................................................................................S ..N...Y.S..S.TP.....HY.YG.S..T....Q..SSTCTG............T......V.....NX CRY1_bosTau .............................................................................I....... .................................................................................................S ..N...C.SNAS.TQ.....HY..G.S..T....Q..SST.AG.G..........T......V.....NX CRY1_equCab .............................................................................I................................................................................................................................................S ..N...C.S..S.SQ.....HY..G.S..T....Q..SSL..G.......GP...T.G....V.....TX CRY1_canFam .............................................................................I................................................................................................................................................S ..N...C.S..S.SQ.....HY..G.S..T....Q..SSM.TG........E...T.T.S..V.....NX CRY1_myoLuc ......................................................................V......I.................................................................A..............S...............................................................S ..N...C.S..SY.Q.....HY.LG.S..T....Q..SS..TG............T....R.V.....NX CRY1_pteVam .............................................................................I................................................................................................................................................S ..N...C.S..S.SQ...S.HY..G.C..T....Q..SSM.TG............M......V.....NX CRY1_eriEur .............................................................................I..............................................................................................................................................A.S ..N..VC.S..S.SQ.....HY..G.S..T....Q..SS..TGF.N.........M. ..V...C.NX CRY1_sorAra .............................................................................I.................................................................................................................................T..........F...S ..NAS.C.S..SYFQ.....RY.YG.S..T....Q..SSSCTG............R. ..V...T.NX CRY1_loxAfr .............................................................................I................................................................................................................................................S ..NT..CNS..S.SQ.....HYV.G.S ..Q..SPT.TGV........D.ET.TL...V.....NX CRY1_proCap .............................................................................I....... ..................... ...............................................................................................................I..S ......C.N..S.SQ.....HY..G.S..........SPM.TGI..........ET.TV.R.V.....NX CRY1_echTel .........S........S..........................................................I................................................................................................................................................S ..NTT.C.SG...PP.N...HY..G.S...A...Q..SPL.TG............T..V...V....SNX CRY1_dasNov .............................................................................I................................................................................................................................................. ..N.L.C.S..S..Q..S..HY..G.N..T....Q..SSM.T............ET......V.....NX CRY1_choHof .....T................H....N...T.T...K.....................L..SI........P....I....... ..............................N.. ......S...............................................................................................S ..N...C.S.. .SQ.....HY..G.S..T....Q..SSM.IG...........ET.G....V.....NX
The place of mole-rats within rodent phylogeny
The issue here is how long has the selective pressure been relaxed on naked mole-rat opsins. For that, close-in outgroups and a dated phylogenetic tree are needed. Seven other rodents have genome projects available so opsins can be extracted to obtain a baseline for mole-rat opsins and unusual properties they might have. Note guinea pig is the immediate outgroup to mole-rat among genome projects, not mouse or rat.
Unfortunately no genomes have been completed for species with convergent life histories such as blind mole rat (Spalax ehrenbergi) insectivore moles, African golden moles, or marsupial moles. Individual opsins are sometimes available for these species, importantly LWS, melanopsin and rhodopsin for middle eastern blind mole rat (a sciuriomorph rodent), rhodopsin fragments for insectivore moles, and short wavelength opsin fragments in a related genus of naked mole rat.
Thus for most opsins, the only available control sequences are from guinea pig, a long-domesticated hystricognath rodent from South America most closely related to a local crepuscular grassland wild species. Since the subterranean naked-mole rat is from East Africa and the two continents separated 103 million years ago, the two species cannot be too closely related. The hystricognath rodents separated from sciuromorphs some 69 million years ago. Based on fossil evidence the hystricognath ancestor of guinea pig arrived in South America by tree raft around 30 million years ago (early Oligocene).
The recently discovered Laotian rock-rat was initially thought to be a hystricognath rodent, which would represent a great geographic oddity. However molecular data places it as the immediate outgroup. The zygomasseteric system of jaw muscle insertion anatomy is not an attractive taxonomic character today. Rare genomic events such as retroposon insertion support this positioning of rock-rat as well as a more nuanced sistering of ctenohystricognaths to murids to the exclusion of the squirrel clade.
Consistent with this, the most recent molecular evidence places the divergence of Bathyerigidae (mole-rat) and Cavidae (guinea pig) at 47.2 million years (average of eight methods, supplemental Table S7a of Sept 2011 Science). This is not an ideal outgroup situation for assessing opsin change attributable to subterranean lifestyle -- that time frame is tantamount to comparing skunks and walruses.
Note the oldest known hystricognath rodent fossil, found recently in Egypt, puts the split between phiomorphs and caviomorphs quite a bit earlier, at 36 million years
Two options here for better mole-rat control species are Petromus typicus (dassie-rat) which lives above ground in rocky outcrops (lifespan unknown but probably short) and Thryonomys gregorianus (cane rat) a larger agricultural pest with lifespan reported as 5.4 years. A rhodopsin fragment is available for cane rat and utilized in the rhodopsin comparison.
Rhodopsins are also available for the even more closely related Bathyergidae but these have similar underground habits as mole-rat and seemingly similarly limited vision despite apparently normal rhodopsins -- note only Fukomys is eusocial; it like the solitary Heliophobius and Bathyergus has full hair coats).
The rate of rhodopsin evolution would not only differs between diurnal and subterranean rodents but also differs because of different tempo and mode overall of rodent and primate evolution. This highlights the need for close-in outgroups for all opsin comparisons
Without a series of genomic controls, the decay of visual apparatus in mole-rat cannot really be dated though obviously onset of strict subterranean life is the most plausible beginning. For onset of longevity, genomes of closely related shorter-lived species are probably essential.
Reference sequences
The sequences below are hand-curated directly from genome project and transcript data, an imperative because many GenBank gene entries are complete rubbish. In the case of mole-rat, orthologous sequences from other hystricognath rodents are very important controls, as are sequences from other subterranean or cave vertebrates.
Murid rodents have been better studied and so are important for PubMed-based annotation transfer; however apart from mouse, genomic coverage is quite poor. The same can be said for lagomorphs. Here small contigs allow ortholog recovery only on an exon-by-exon basis. That is slightly problematic for genes having little divergence because a paralogous exon might be confused with the authentic orthologous exon.
Some observed oddities are genuine; others may be sequencing or assembly errors. In some cases, that can resolved by reviewing raw read data, by transcript support, by on-target publications, or by comparative genomic considerations in lieu of experimental re-sequencing.
Much larger collections of curated metazoan opsins and cryptochromes are available elsewhere.
Rodent opsins
The comparative genomics of opsins have been studied exhaustively: no variation in exon number or intron location or phasing occurs in any terrestrial vertebrate. The sequences below have one exon per row. The numbers 0 1 2 indicate the observed codon overhang (reading frame). These are deeply conserved within eumetazoans; mole-rat presents no new issues. Processed pseudogenes are absent in opsins, consistent with no gene expression of opsins in terminal germline cell lineages.
>RHO1_hetGla Heterocephalus glaber (naked molerat) RHO syn(-MBD4 +IFT122 +H1FOO -PLXND1) EHB14304
0 MNGTEGPNFYVPFSNITGMVRSPFEYPQYYLAEPWQFSMLAAYMFLLIVLGFPVNFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVICGFTTTLYTSMHGYFVFGATGCNMEGFFATLG 1
2 GEIALWSLVVLAIERYMVVCKPMSNFRFGENHAIVGVAFTWVMALACAAPPLAGWSR 2
1 YIPEGMQCSCGIDYYTPKPEINNESFVIYMFVVHFTIPLVIIFFCYGQLVFTVKE 0
0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWVPYASVAMYIFTHQGSDFGPIFMTIPAFFAKSSSIYNPVIYIMMNKQ 0
0 FRNCMLTTICCGKSPLGDDEASTTASKTETSQVAPA* 0
>LWS_hetGla Heterocephalus glaber (naked molerat) OPN1LW syn(-IRAK1 -MECP2 -TEX28) 88% identical musMus, missing exons 1-5 because of assembly gaps
0 1
2 1
2 2
1 2
0 0
0 FPNCILQLFGKKVEDSLELSSASKTEASSVSSVSPA* 0
>SWS1_hetGla Heterocephalus glaber (naked molerat) OPN1SW syn(-FAM137A +SWS1 -CALU -NAG6 -FLNC) 89% identical musMus, frameshifting deletion 6aa exon 4
0 MSGEEDFYLFKNTSSVGPWDGSQYHIAPIWAFHLQAAFMGLVFFVGTPLNAIVLVATLQYKKLRQPLNYILVNVSLGGFLFCIFSVFTVFIASCHGYFFFGRHVCALEAFLGSVA 1
2 GLVTGWSLAFLAFERYLVICKPFGNFRFSSKHALIVVLATWVIGIGVSIPPFFGWSR 2
1 YIPEGLQCSCGPDWYTVGTKYRSEYYSWFLFIFCFIVPLSLICFSYSQLLRTLRA 0
0 VAAQQQESATTQKAEREVSRMVVVMVGSFCVCYVPYAALAMYIVNNRNHGLDLRLFTIPAFFSPKSCVYNPIIYCFMNKQ 0
0 FRACIMELVCRKSMADESDMSSSQKTEVSAVSSSKVGPN* 0
>MEL1_hetGla Heterocephalus glaber (naked molerat) OPN4 melanopsin AFSB01020105
0 MDTPSGPRVSPDPAQEPSFLATLSAPGRGDGTPSSLSTPGQLPPGNPT 0
0 AAGTQAAVRVPFSTADVPAHVHYTLGAVILLVGLTGMLGNLTVIYTFFR 2
1 SRGLRTPANMFVINLAVSDFLMSFTQAPVFFTSSLYKRWLFGEA 2
2 GCEFYAFCGALFGITSMITLTAITLDRYLVITRPLATIGLASKRQAALVLLGIWLYALAWSLPPFFGW 1
2 sAYVPEGLLTSCSWDYMTFTPSVRSYTMLLFCFVFFLPLLIIIYCYVFIFRAIRETGR 2
1 AFESCGESPSRRRQWQRLRSEWKMAKIALLVILLFVLSWAPYSTVALVAFAG 2
1 HAHILTPSMSSVPAVIAKASAIHNPIIYAIAHPKYR 2
1 MAIVQHLPCLGLLLGVSGQRSLPSLSYRSTHRSRLSSQASDLSWISGRRRQESLGSESEV 0
0 GWTDTEAWGSAQQASERPPCGTGLEGWEARAHPKAQEWEAETPGK 0
0 TKGRLPSLDPQM* 0
>ENC_hetGla Heterocephalus glaber (naked molerat) OPN3 88% identical musMus, missing most of first exon AFSB01146037
0 VSCLRNGWVWDAVGCVWDGFSSSLF 1
2 GIVSITTLTVLAYERYIRVVHARVINFSWAWKAITYIWLYSLAWAGAPLLGWNRYILDVHGLSCTVDWKSKDANDSSFVLFLFLGCLVVPMGVIAHCYGHILYSIRM 0
0 FSMLQLRCVEDLQTIQVMKILRYEKKVAKMCFLMVFIFLVCWMPYIVICFLVVNGYGHRVTPTVSVVSYLFAKSSTVYNPVIYTIMIRK 0
0 FRRSLLQLLCFRLLRCQQPAKDLPAVESEMQIRPIVMSQKDGDRPKKKVTFNSSSIIFIITSDESLSVDDSDRTSGSKVDTIQVRPL* 0
>NEUR1_hetGla Heterocephalus glaber (naked molerat) OPN5 94% identical musMus
0 MALNHTAPPQDERLPHYLQDEDPFVSKLSWEADLVAGFYLTII 1
2 GILSTCGNGYVLYMSSRRKKKLRPAEIMTINLAICDLGIS 1
2 VVGKPFTIISCFRHRWVFGWIGCRWYGWAGFFFGCGSLTTMTVVSLDRYLKICYLSY 1
2 GVWLKRKHAYICLAAIWAYVSFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASARGQVFILNILFFCLLLPMATIVFSYAKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTK 0
0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYRFACCQNGGLKATKKKSLEDFR 2
1 LHTVTTVRKSSAVLEIHQEV* 0
>PER_hetGla Heterocephalus glaber (naked molerat) RRH syn(-CFI +NOLA1 +EGF -ELOVL6) AFSB01025324 missing exon 7
0 MLRNSFGNGSDSKSEDGSVFSRMEHSIVAAWLLLA 1
2 GLISILSNIIVLGIFVKYKELRTPTNAIIMNLALTDIGVSSIGYPMSAASYLHGSWKFGYPGCQ 0
0 VYAGLNIFFGMASIGLLTVIAVDRYLTICRPDI 1
2 GRRLTSTSYVSMILGAWINGLFWALMPIIGWASYAPDPTGATCTINWRKNDt 2
1 SFVSYTMTVIAVNFTGPLAVMFYCYFHVTWSIKHHATGNCPTFPNRDWSDQVDVTK 0
0 MSVVMILMFLVAWSPYSIVCLWASFGDPKKIPPSMAIIAPLFAKSSTFYNPCIYVIANKK 2
1 FRRAMCTMFRCQTHQAMPETSALPMGASQSPLASG* 0
>RGR_hetGla Heterocephalus glaber (naked molerat) RGR syn(+PCDH21 -LRIT1 -GRID1 -WAPAL) 74% identical musMus, missing exons 1
0 MAASGALPTGFGELEVLAVGTMLLVE 1
2 ALCGLSLNGLTIVSFCKIPELWTPRHLLVLSLALADSGLSLNTLIAAVSSLLR 2
1 RWPYGSGGCQTHGFQGFVTALASISGSAAIAWGHHQHYCT 1
2 GSQLAWSTAIRLVLFVWLSSAFWAALPLLGWGHYDYEPLGTCCTLDYSKRDR 2
1 NSTSFLFTMGFFSFLIPLFITFTSYQLMEQKLARSSRLQ 0
0 VNTSLPARTLMLCWGPYVFLHLYAVIADASSLSPKLQM 0
0 VPALIAKTVPTINAINYALGSKMGSRRPWQCLSLQRSKRD* 0
>RHO1_helArt Heliophobius argenteocinereus (silvery mole-rat) solitary, furred, weak eyes
0 VPFSNITGVVRSPFEYPQYYLAEPWQFSMLAAYMFLLIMLGFPVNFLTLYVTVQHKKLRTPLNYILLNLAVANLFMVICGFTTTLYTSMHGYFVFGPTGCNMEGFFATLG 1
2 GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVTFTWVMALACAAPPLAGWSR 2
1 YIPEGMQCSCGIDYYTPKPEINNESFVIYMFVVHFTIPLVIIFFCYGQLVFTVKE 0
0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWVPYASVAVYIFTHQGSDFGPIFMTIPSFFAKSSSIYNPVIYIMMNKQ 0
0 FRNCMLTTICCGKNPLGDDEAYTTAS * 0
>RHO1_cryDam Cryptomys damarensis (damarkind mole-rat) GQ290302 genus changed to Fukomys
0 VPFSNITGVVRSPFEYPQYYLAEPWQFSVLAAYMFLLIVLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVANLFMVICGFTTTLYTSMHGYFVFGPTGCNMEGFFATLG 1
2 GEIALWSLVVLAIERYMVVCKPMSNFRFGENHAVMGVAFTWVMALACAAPPLAGWSR 2
1 YIPEGMQCSCGIDYYTPKPEINNESFVIYMFVVHFTIPLVIIFFCYGQLVFTVKE 0
0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWVPYASVAVYIFTHQGSDFGPIFMTIPAFFAKSSSIYNPVIYIMMNKQ 0
0 FRNCMLTTICCGKNPLGDDEASTTAS * 0
>RHO1_batSui Bathyergus suillus (dune mole-rat)
0 VPFSNITGVVRSPFEYPQFYLAEPWQFSMLAAYMFLLIVLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVICGFTTTLYTSMHGYFIFGPTGCNMEGFFATLG 1
2 GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVALTWVMALACAAPPLAGWSR 2
1 YIPEGMQCSCGIDYYTPKPEINNESFVIYMFVVHFTIPLVIIFFCYGQLVFTVKE 0
0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWVPYASVAVYIFTHQGSDFGPIFMTIPAFFAKSSSIYNPVIYIMMNKQ 0
0 FRNCMLTTICCGKNPLGDDEASTTAS * 0
>RHO1_thySwi Thryonomys swinderianus (cane-rat) GQ290319
0 VPFSNATGVVRSPFEYPQYYLAEPWQFSMLAAYMFLLIVLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLG 1
2 GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLAGWSR 2
1 YIPEGMQCSCGIDYYTLKPEVNNESFVIYMFVVHFTIPMIIIFFCYGQLVFTVKE 0
0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWVPYASVAMYIFTHQGSNFGPIFMTLPAFFAKSASIYNPVIYIMMNKQ 0
0 FRNCMLTTICCGKNPLGDDEASTTAS * 0
>RHO1_cavPor Cavia porcellus (guinea_pig)
0 MNGTEGENFYIPFSNATGVVRSPFEYPQYYLAEPWQFSILAAYMFMLIVLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVANLFMVLGGFTTTLYTSMNGYFVFGPTGCNLEGFFATLG 1
2 GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVVFTWIMALACAAPPLVGWSR 2
1 YIPEGMQCSCGVDYYTLKSEVNHESFVIYMFVVYFTIPMIIIFFCYEQLVFTVKE 0
0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWVPYASVAAYIFTHQGSNFGPIFMTVPAFFAKSSSIYNPVIYIMMNKQ 0
0 FRNCMLTTICCGKNPLGDDEASTTVSKTETSQVAPA* 0
>RHO1_musMus Mus musculus (mouse) RHO syn(-MBD4 +IFT122 +H1FOO -PLXND1)
0 MNGTEGPNFYVPFSNVTGVVRSPFEQPQYYLAEPWQFSMLAAYMFLLIVLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLG 1
2 GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVVFTWIMALACAAPPLVGWSR 2
1 YIPEGMQCSCGIDYYTLKPEVNNESFVIYMFVVHFTIPMIVIFFCYGQLVFTVKE 0
0 AAAQQQESATTQKAEKEVTRMVIIMVIFFLICWLPYASVAFYIFTHQGSNFGPIFMTLPAFFAKSSSIYNPVIYIMLNKQ 0
0 FRNCMLTTLCCGKNPLGDDDASATASKTETSQVAPA* 0
>RHO1_ratNor Rattus norvegicus (rat)
0 MNGTEGPNFYVPFSNITGVVRSPFEQPQYYLAEPWQFSMLAAYMFLLIVLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLG 1
2 GEIGLWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLVGWSR 2
1 YIPEGMQCSCGIDYYTLKPEVNNESFVIYMFVVHFTIPMIVIFFCYGQLVFTVKE 0
0 AAAQQQESATTQKAEKEVTRMVIIMVIFFLICWLPYASVAMYIFTHQGSNFGPIFMTLPAFFAKTASIYNPIIYIMMNKQ 0
0 FRNCMLTTLCCGKNPLGDDEASATASKTETSQVAPA* 0
>RHO1_nanEhr Nannospalax ehrenbergi (mole-rat) AAG25707
MNGTEGPNFYVPFSNGTGVVRSPFEQPQYYLAEPWQFSMLAAYMFLLIVLGFPINFLTLYVTVQHKKLRTPLNYILLNLALADLFMVIGGFTTTFYTSLHGYFVFGPTGCNLEGFFATLG 1
2 GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAITGVAFTWVMALACAVPPLVGWSR 2
1 YIPEGMQCSCGIDYYTLKPEVNNESFVIYMFVVHFTIPMIIIFFCYGQLVFTVKE 0
0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWVPYASVAMYIFTHQGSNFGPIFMTLPAFFAKSASIYNPVIYIMMNKQ 0
0 FRNCMLTTLCCGKNLLGDEEASPTASKTETSQVAPA* 0
>RHO1_criGri Cricetulus griseus (hamster) EGW12630
MNGTEGPNFYVPFSNATGVVRSPFEYPQYYLAEPWQFSMLAAYMFLLIVLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLG 1
2 GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVVFTWIMALACAAPPLVGWSR 2
1 YIPEGMQCSCGVDYYTLKPEVNNESFVIYMFVVHFTIPLIVIFFCYGQLVFTVKE 0
0 AAAQQQESATTQKAEKEVTRMVILMVVFFLICWVPYAGVAFYIFTHQGSNFGPIFMTLPAFFAKSSSIYNPVIYIMMNKQ 0
0 FRNCMLTTLCCGKNILGDDEASATASKTETSQVAPA* 0
>RHO1_oryCun Oryctolagus cuniculus (rabbit)
MNGTEGPDFYIPMSNQTGVVRSPFEYPQYYLAEPWQFSMLAAYMFLLIVLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVLGGFTTTLYTSLHGYFVFGPTGCNVEGFFATLG 1
2 GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWIMALACAAPPLVGWSR 2
1 YIPEGMQCSCGIDYYTLKPEVNNESFVIYMFVVHFTIPLIIIFFCYGQLVFTVKE 0
0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWVPYASVAFYIFTHQGSNFGPIFMTIPAFFAKSSSIYNPVIYIMMNKQ 0
0 FRNCMLTTICCGKNPLGDDEASATASKTETSQVAPA* 0
>LWS_cavPor Cavia porcellus (guinea_pig)
0 MAQRWGPHALSGVQAQDAYEDSTQASLFTYTNSNNTR 1
2 GPFEGPNYHIAPRWVYHLTSAWMTIVVIASIFTNGLVLVATMRFKKLRHPLNWILVNLAVADLAETVIASTISVVNQVYGYFVLGHPLCVVEGYTVSLC 1
2 GITGLWSLAIISWERWLVVCKPFGNVRFDAKLAIVGIVFSWVWSAVWTAPPIFGWSR 2
1 YWPYGLKTSCGPDVFSGTSYPGVQSYMMVLMVTCCITPLSIIVLCYLHVWLAIRA 0
0 VAKQQKESESTQKAEKEVTRMVVVMVLAYCLCWGPYAFFACFATANPGYSFHPLVAALPAYFAKSATIYNPIIYVFMNRQ 0
0 FRNCILQLFGKKVEDSSELSSTSRTEASSVSSVSPA* 0
>LWS_musMus Mus musculus (mouse) Opn1mw = OPN1LW syn(-IRAK1 -MECP2 -TEX28)
0 MAQRLTGEQTLDHYEDSTHASIFTYTNSNSTK 1
2 GPFEGPNYHIAPRWVYHLTSTWMILVVVASVFTNGLVLAATMRFKKLRHPLNWILVNLAVADLAETIIASTISVVNQIYGYFVLGHPLCVIEGYIVSLC 1
2 GITGLWSLAIISWERWLVVCKPFGNVRFDAKLATVGIVFSWVWAAIWTAPPIFGWSR 0
0 YWPYGLKTSCGPDVFSGTSYPGVQSYMMVLMVTCCIFPLSIIVLCYLQVWLAIRA 0
0 VAKQQKESESTQKAEKEVTRMVVVMVFAYCLCWGPYTFFACFATAHPGYAFHPLVASLPSYFAKSATIYNPIIYVFMNRQ 0
0 FRNCILHLFGKKVDDSSELSSTSKTEVSSVSSVSPA* 0
>LWS_ratNor Rattus norvegicus (rat)
0 MAQQLTGEQTLDHYEDSTQASIFTYTNSNSTR 1
2 GPFEGPNYHIAPRWVYHLTSTWMILVVIASVFTNGLVLAATMRFKKLRHPLNWILVNLAVADLAETIIASTISVVNQIYGYFVLGHPLCVIEGYIVSLC 1
2 GITGLWSLAIISWERWLVVCKPFGNVRFDAKLATVGIVFSWVWAAVWTAPPIFGWSR 2
1 YWPYGLKTSCGPDVFSGTSYPGVQSYMMVLMVTCCIFPLSIIVLCYLQVWLAIRA 0
0 VAKQQKESESTQKAEKEVTRMVVVMVFAYCLCWGPYTFFACFATAHPGYAFHPLVASLPSYFAKSATIYNPIIYVFMNRQ 0
0 FRNCILQLFGKKVDDSSELSSTSKTEVSSVSSVSPA* 0
>LWS_nanEhr Nannospalax ehrenbergi (mole_rat)
0 MAQQWAPQRLAGGQTQDSYEDSTQASLFTYTNSNSTR 1
2 GPFEGPNYHIAPRWVYHLTTTWMILVVVASIFTNGLVLVATMRFKKLRHPLNWILVNLAVADLAETIIASTISVVNQIYGYFVLGHPLCVIEGYTVSLC 1
2 GITGLWSLAIISWERWMVVCKPFGNVRFDAKLATMGITFAWVWAAVWTAPPVFGWSR 2
1 YWPYGLKTSCGPDVFSGTSYPGVQSYMVVLMITCCIIPLSIILLCYLQVWLAIRT 2
0 VAKQQKESESTQKAEKEVTHMVVVMVFAYCLCWGPYTFFVCFATAHPGYAFHPLVAALPAYFAKSATIYNPIIYVFMNRQ 0
0 FQNCILQLFGKKVDDSSELSSTSKTEASSVSSVSPA* 0
>LWS_speTri Spermophilus tridecemlineatus (squirrel) Ictidomys
0 MAQRWDPQRLAGGQPQDSHEDSTQSSIFTYTNSNATR 1
2 GPFEGPNYHIAPRWVYHITSIWMIIVVIASVFTNGLVLVATMKFKKLRHPLNWILVNLAIADLAETVIASTISVVNQFFGYFVLGHPLCVVEGYTVSVC 1
2 GITGLWSLAIISWERWLVVCKPFGNVRFDAKLAIVGIAFAWTWSAVWTAPPIFGWSR 2
1 YWPYGLKTSCGPDVFSGNSYPGVQSYMMVLMVTCCIIPLSIIILCYLQVWLAIRA 0
0 VAKQQKESESTQKAEKEVTRMVVVMVLAYCLCWGPYASFACFATANPGYAFHPLVAAVPAYFAKSATIYNPVIYVFMNRQ 0
0 FRNCILQLFGKKVDDTSELSSASRTEASSVSSVSPA* 0
>LWS_sciCar Sciurus carolinensis (squirrel) frag
0 mAQRWDPQRLAGGQPQDSHEDSTQSSIFTYTNSNATR 1
2 GPFEGPNYHIAPRWVYHITSTWMIIVVIASVFTNGLVLVATMKFKKLRHPLNWILVNLAIADLAETVIASTISVVNQLYGYFVLGHPLCVVEGYTVSVC 1
2 GITGLWSLAIISWERWLVVCKPFGNMRFDAKLAIVGIAFSWIWSAVWTAPPIFGWSR 2
1 YWPYGLKTSCGPDVFSGTSYPGMQSYMMVLMVTCCIIPLSIIILCYLQVWLAIRA 0
0 VAKQQKESESTQKAEKEVTRMVVVMVFAYCLCWGPYTFFACFATANPGYAFHPLVAALPAYFAKSATIYNPIyvfmnrq 0
0 * 0
>LWS_oryCun Oryctolagus cuniculus (rabbit)
0 MTQPWGPQMLAGGQPPESHEDSTQASIFTYTNSNSTR 1
2 GPFEGPNFHIAPRWVYHLTSAWMILVVIASVFTNGLVLVATMRFKKLRHPLNWILVNLAVADLAETVIASTISVVNQFYGYFVLGHPLCVVEGYTVSLC 1
2 GITGLWSLAIISWERWLVVCKPFGNVRFDAKLAIAGIAFSWIWAAVWTAPPIFGWSR 2
1 YWPYGLKTSCGPDVFSGTSYPGVQSYMMVLMVTCCIIPLSVIVLCYLQVWMAIPA 0
0 VAKQQKESESTQKAEKEVTRMVVVMVFAYCLCWGPYTFFACFATAHPGYSFHPLVAAIPSYFAKSATIYNPIIYVFMNRQ 0
0 FRNCILQLFGKKVEDSSELSSASRTEASSVSSVSPA* 0
>SWS1_cavPor Cavia porcellus (guinea_pig) Hystricognathi
0 MSEEEDFYLFKNASSVGPWDGPQYHVAPVWAFRLQAAFMGIVFCIGTPLNGIVLVATLLYKKLRQPLNYILVNVSLGGFLVCIFS
MVTGWSLAFLAFERYLVICKPFGNFRFSSKHALIVVLATWVIGIGVSIPPFFGWSR 2
1 YMPEGLQCSCGPDWYTMGTKYRSEYFAWFLFIFCFIVPLSLICFSYCQLLRTLRT 0
0 VAAQQQESATTQKAEREVSRMVVVMVGSFCVCYVPYAALAMYIVNNRNHGLDLRLVTIPAFFSKSSCIYNPIIYCFMNKQ 0
0 FRACIMELVCRKPMADESDMSTSQKTEVSAVSSSKVGPN*
>SWS1_cteTal Ctenomys talarum (tuco-tuco) ADF36522 Hystricognathi
LQASFMGFVFFAGTPLNAIVLVATLKYKKLRQPLNYILVNVSLGGFLFCVFSVSTVFIASCHGYFLFGREVCALEAFLGSVA
>SWS1_cteMag Ctenomys magellanicus (tuco-tuco) ADF36521 Hystricognathi
LQASFMGFVFFAGTPLNAIVLVATLKYKKLRQPLNYILVNVSLGGFLFCVFSVSTVFIASCHGYFLFGREVCALEAFLGSVAGLVTGWSLAFLAFERYLVICKPFGNFRFSSKHALMVVLATWVIGIGVSIPPFFGWSRYIPEGLQCSCGP
>SWS1_spaCyn Spalacopus cyanus (coruro) ADF36520 Hystricognathi
LQAAFMGFVFFAGTPLNAIVLVATLQYKKLRQPLNYILVNVSLGGFLFCIFSVFTVFIASCHGYFLFGHQVCALEAFLGSVAGLVTGWSLAFLALERYLVICKPFGNFRFSSKHALMVVLATWVIGIGVSIPPFFGWSRYIPEGLQCSCGP
>SWS1_octDeg Octodon degus (degu) ADF36519 Hystricognathi
LQAAFMGFVFFAGTPLNAIVLVATLQYKKLRQPLNYILVNVSLGGFLFCIFSVFTVFIASCHGYFLFGHQVCALEAFLGSVAGLVTGWSLAFLALERYLVICKPFGNFRFSSKHALMVVLATWVIGIGVSIPPFFGWSRYIPEGLQ
>SWS1_talEur Talpa europaea (mole) ABV31135 Laurasiathere Insectivora
FAGTPLNATVLVATLRYRKLRQPLNYILVNVSLGGFLFCIFSVLTVFIASCKGYFIFGRHVCALEAFLGSAAGLVTGWSLAFLAFERYIVICKPFGNFR
>SWS1_musMus Mus musculus (mouse) OPN1SW syn(-FAM137A +SWS1 -CALU -NAG6 -FLNC)
0 MSGEDDFYLFQNISSVGPWDGPQYHLAPVWAFRLQAAFMGFVFFVGTPLNAIVLVATLHYKKLRQPLNYILVNVSLGGFLFCIFSVFTVFIASCHGYFLFGRHVCALEAFLGSVA 1
2 GLVTGWSLAFLAFERYVVICKPFGSIRFNSKHALMVVLATWIIGIGVSIPPFFGWSR 2
1 FIPEGLQCSCGPDWYTVGTKYRSEYYTWFLFIFCFIIPLSLICFSYSQLLRTLRA 0
0 VAAQQQESATTQKAEREVSHMVVVMVGSFCLCYVPYAALAMYMVNNRNHGLDLRLVTIPAFFSKSSCVYNPIIYCFMNKQ 0
0 FRACILEMVCRKPMADESDVSGSQKTEVSTVSSSKVGPH* 0
>SWS1_ratNor Rattus norvegicus (rat)
0 MSGEDEFYLFQNISSVGPWDGPQYHIAPVWAFHLQAAFMGFVFFAGTPLNATVLVATLHYKKLRQPLNYILVNVSLGGFLFCIFSVFTVFIASCHGYFLFGRHVCALEAFLGSVA 1
2 GLVTGWSLAFLAFERYLVICKPFGNIRFNSKHALTVVLITWTIGIGVSIPPFFGWSR 2
1 FIPEGLQCSCGPDWYTVGTKYRSEHYTWFLFIFCFIIPLSLICFSYFQLLRTLRA 0
0 VAAQQQESATTQKAEREVSHMVVVMVGSFCLCYVPYAALAMYMVNNRNHGLDLRLVTIPAFFSKSSCVYNPIIYCFMNKQ 0
0 FRACILEMVCRKPMTDESDMSGSQKTEVSTVSSSKVGPH
>SWS1_criGri Cricetulus griseus (hamster) XP_003497566
0 MSGEDEFYLFKNISSVGPWDGPQYHIAPVWAFHLQAAFMGFVFLAGTPLNATVLVVTLRYKKLRQPLNYILVNVSLGGFLFCSFSVFTVFIASCHGYFLFGRHVCALEAFLGSVA 1
2 GLVTGWSLAFLAFERYIVICKPFGNIRFSSKHALMVVLVTWTIGIGVSIPPFFGWSR 2
1 FIPEGLQCSCGPDWYTVGTKYRSEYYTWFLFLFCFIVPLSLICFSYSQLLRTLRA 0
0 VAAQQQESATTQKAEREVSHMVVVMVGSFCLCYVPYAALAMYMVNNRNHGLDLRLVTIPAFFSKSSCVYNPIIYCFMNKQ 0
0 FRACIMEMVCRKPVTDESDMSGSQKTEASTVSSSKVGPH* 0
>MEL1_cavPor Cavia porcellus (guinea pig) AAKN02044549
0 MEPSSGPTQGSSVSATLPAPSRGDDTLSSLFTPAPLPPGSPT 0
0 VAGAQAAAWIPFPTVDVPAHAHYTLGAVILLVGLTGMLGNLTVIYTFCR 2
1 SRGLRTPANMFIINLAVSDFLMSFTQAPVFFTSSLYKRWLFGEA 2
2 GCEFYAFCGALFGITSMITLTAITLDRYLVITRPLATIGVASKRQAALVLLGVWLYALAWSLPPFFGW 1
2 SAYVPEGLLTSCSWDYVTFTPSVRAYTMLLFCFVFFLPLLVIIYCYIFIFRAIRETGR 2
1 AFESCGESPRRRRQWQRLRSEWKMAKIALLVILLFVLSWAPYSAVALVAFAG 2
1 YAHMLTPYMNSVPAVIAKASAIHNPIIYAITHPKY 2
1 AAIVQHLPCLGVLLGVSGQHGRPSLRYRSTHRSTLSSQASDLSWISGRRRQESLGSESEV 0
0 GWTDTEAWGAAQQGSGQSPYDLVLEDWEARAPPKARVWGAGTPGK 0
0 TKGQLTCLDPQM* 0
>MEL1_musMus Mus musculus (mouse) OPN4 melanopsin pentultimate exon sometimes skips 6 residues
0 MDSPSGPRVLSSLTQDPSFTTSPALQGIWNGTQNVSVRAQLLSVSPT 0
0 TSAHQAAAWVPFPTVDVPDHAHYTLGTVILLVGLTGMLGNLTVIYTFCR 2
1 NRGLRTPANMFIINLAVSDFLMSVTQAPVFFASSLYKKWLFGET 1
2 GCEFYAFCGAVFGITSMITLTAIAMDRYLVITRPLATIGRGSKRRTALVLLGVWLYALAWSLPPFFGW 1
2 SAYVPEGLLTSCSWDYMTFTPQVRAYTMLLFCFVFFLPLLIIIFCYIFIFRAIRETGR 2
1 ACEGCGESPLRQRRQWQRLQSEWKMAKVALIVILLFVLSWAPYSTVALVAFAG 2
1 YSHILTPYMSSVPAVIAKASAIHNPIIYAITHPKYR 2
1 VAIAQHLPCLGVLLGVSGQRSHPSLSYRSTHRSTLSSQSSDLSWISGRKRQESLGSESEV 0
0 GWTDTETTAAWGAAQQASGQSFCSQNLEDGELKASSSPQVQRSKTPK 0
0 TKGHLPSLDLGM* 0
>MEL1_ratNor Rattus norvegicus (rat) AY072689
0 MNSPSESRVPSSLTQDPSFTASPALLQGIWNSTQNISVRVQLLSVSPT 0
0 TPGLQAAAWVPFPTVDVPDHAHYTLGTVILLVGLTGMLGNLTVIYTFCR 2
1 NRGLRTPANMLIINLAVSDFLMSFTQAPVFFASSLYKKWLFGET 1
2 GCKFYAFCGAVFGIVSMITLTAIAMDRYLVITRPLATIGMRSKRRTALVLLGVWLYALAWSLPPFFGW 1
2 SAYVPEGLLTSCSWDYVTFTPLVRAYTMLLFCFVFFLPLLIIIFCYIFIFRAIRETGR 2
1 ACEGCGESPLRRRQWQRLQSEWKMAKVALIVILLFVLSWAPYSTVALVGFAG 2
1 YSHILTPYMSSVPAVIAKASAIHNPIIYAITHPKYR 2
1 AAIAQHLPCLGVLLGVSGQRSHPSLSYRSTHRSTLSSQSSDLSWISGQKRQESLGSESEV 0
0 GWTDTETTAAWGAAQQASGQSFCSHDLEDGEVKAPSSPQEQKSKTPK 0
0 TKRHLPSLDRRM* 0
>MEL1_criGri Cricetulus griseus (hamster) AFTD01055861 XM_003495335 mssing last exon
0 MDSPSGPTGPPGLTQGPSFMASTALQGHWNSTQKVSTRVQLLSMSPT 0
0 ASGPEAAAWVPFPTVDVPDHAHYTLGTVILLVGLTGMLGNLTVIYTFCR 2
1 NRGLRTPANMFIINLAVSDFLMSFTQAPVFFASSLYKKWLFGET 1
2 GCEFYAFCGAVFGITSMITLTAIALDRYLVITRPLATIGMGSKRRTAFVLLGVWLYALAWSLPPFFGW 1
2 SAYVPEGLLTSCSWDYMTFTPQVRAYTMLLFCFVFFLPLLIIIGCYIFIFRAIRETGR 2
1 ACEGCSESPQQRRQWHRLQSEWKMAKVALIVILLFVLSWAPYSTVALVAFAG 2
1 YSHILTPYMSSVPAVIAKASAIHNPIIYAITHPKYR 2
1 AAIAQHLPCLGVLLGVSSQRSHPSLSYRSTQRSTLSSQSSDLSWISGPKRQESLGSESEV 0
0 GWTETEATAVWGTAQPASGQSSCSQNLQDGAVKAPSSPQVQNSKAPK 0
0 AKGQLPSLDLGM* 0
>MEL1_phoSun Phodopus sungorus (hamster) AY726733 short form amended with vqnskapk and run-out QDAP* dropped
0 MDSPPGPTAPPGLTQGPSFMASTTLHSHWNSTQKVSTRAQLLAVSPT 0
0 ASGPEAAAWVPFPTVDVPDHAHYILGTVILLVGLTGMLGNLTVIYTFCR 2
1 SRSLRTPANMLIINLAVSDFLMSFTQAPVFFASSLYKKWLFGET 2
1 GCEFYAFCGAVLGITSMITLTAIALDRYLVITRPLATIGMGSKRRTALVLLGIWLYALAWSLPPFFGW 1
2 SAYVPEGLLTSCSWDYVTFTPQVRAYTMLLFCFVFFLPLLVIIFCYISIFRAIRETGR 2
1 ACEGWSESPQRRRQWHRLQSEWKMAKVALIVILLFVLSWAPYSTVALVAFAG 2
1 YSHILTPYMSSVPAVIAKASAIHNPIVYAITHPKYR 2
1 AAIAQHLPCLGVLLGVSSQRNRPSLSYRSTHRSTLSSQSSDLSWISAPKRQESLGSESEV 0
0 GWTDTEATAVWGAAQPASGQSSCGQNLEDGMVKAPSSPQvqnskapk 0
0 AKGQLPSLDLGM* 0
>MEL1_nanEhr Spalax ehrenbergi (blind molerat) OPN4 AM748539
0 MNSPSGPRVPPGLAQKPSFMVTPVLPNQWISFQKNVSVGIQLPPASAT 0
0 ATGAQAASWVPFPTVDVPVHAHYTLGTVILLVGLTGMLGNLIVIYTFCR 2
1 SRGLRTRANMFTVNLAVSDFLMSFTQAPVFFASSLYKRWLFGEA 2
2 GCEFYAFCGAVSGITSMTTLTAIALDRYLVITRPLATIGVASKRRTALVLLGVWLYALAWSLPPFFGW 1
2 SAYVPEGLLTSCSWDYMTFTPSVRAYTMLLFCFVFFLPLLIIIFCYIFIFKAIRETGR 2
1 ACEGCGESPQRRRQWQRLQNEWKMAKVALLVIFLFVLSWAPYSTVALVAFAG 2
1 YSHILTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2
1 LAISQHLPCLGVLIGVSSQRSHPSLSYRSTHRSTLSSQASDLSWISGRKRQESLGSESEV 0
0 GWTDTEVTAAWGVAQEASGWSPYRHSLEDGEVKASPSPQGQEAKTSRK 0
0 TKGQLPSLNLRM* 0
>MEL1_dipOrd Dipodomys ordii (kangaroo rat) AGTP01108766 ENSDORT00000008232
0 MNPPSRPRISPNPTQEPSFAATPASPGRWDSTQSNVSTPSQLPPISTT 0
0 0
0 TRGLRTPANMFIINLAVSDFLMSFTQAPVFFASSLHKQWLFGEA 1
2 GCEFYAFCGALFGITSMITLMAIALDRYLVITRPLATIGVTSKRRTAFVLLGVWVYALAWSLPPFFGW 1
2 SAYVPEGLLTSCSWDYMTFTPSVRSYTMLLFCFVFFLPLLVIIYCYIFIFRAIRETRR 2
1 ACEGCGESPQQRQQWQRLQSEWKMAKIALLVILLFVLSWAPYSTVALVAFAG 2
1 YAHILTPYMNSVPAVIAKASSIHNPIVYAITHPKYR 2
1 VAIAQHLPCLRVLLGVSGQHDHPSLSFRSTHRSTLTSQTSDLSWISGRRRQESLGSESKM 0
0 GWTDTEAAAAWGDAQQMTGQSPCSQDLEDGEVKVLPRAQRKEAKTPKK 0
0 * 0
>MEL1_ictTri Ictidomys tridecemlineatus (squirrel)
0 MNTTSGSDSPGPAQEPTLGATSAPPSMWDSTQSSVSSLAQLLPASPT 0
0 aTGAQSAARIPFPTVDVPDHAHYTLGTVILLVGLTGLMGNLTVIYTFCR 2
1 SRSLRTPANMFIINLAVSDFLMSFTQAPIFFASSLYKRWLFGEA 2
1 GCEFYAFCGAVFGISSMITLTAIALDRYLVITRPLATIGMASKKRAAFFLLGVWFYALAWSLPPFFGW 1
2 SAYVPEGLLTSCSWDYMTFTPSVRAYTMLLVCFVFFLPLFLIIYCYIFIFRAIRETGR 2
1 ACEGCGESPRRRRQWQRMQSEWKVAKIALLVILLFIVSWAPYSIVALVAFAG 2
1 YAHLLTPYMSSVPAVIAKASAIHNPIIYAIIHPKYR 2
1 MAISQNLPFLKKLLGVSSQHGRPSLSYRSTHRSTLISHASEVSWISGRRRQESLGSESEV 0
0 GWTDTEAAAAWGAAQQMNGWSLCGQILEDEEATAPSRSQGQEAETFRK 0
0 TKGQLPSLCSRM* 0
>MEL1_oryCun Oryctolagus cuniculus (rabbit) ENSOCUT00000017574
0 MNSPWGSRVPPGPAQEPRSTAAPPSRWDGSESSISSPGQLTPGSPT 0
0 APGAQEAAWVPFPTVDVPDHAHYTLGTVILLVGLTGMLGNLTVIYTFCR 2
1 SRSLRTPANMFIINLAVSDFLMSFTQAPVFFASSLYKRWLFGET 2
1 GCEFYAFCGALFGISSMITLTAIALDRYLVITRPLAAVGMVSKKRAGLVLLGVWLYALAWSLPPLFGW 1
2 SAYVPEGLLTSCSWDYVTFTPSVRSYTMLLFCFVFFLPLLVIVYCYIFIFRAIRETGR 2
1 ACQGSHESPQRRQWRRLQSEWKMAKVALLVILLFLLSWAPYSTVALVAFAG 2
1 YAHSLSPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2
1 AAIAQHLPCLGVLLGVSGRHSRPSLSYRSTHRSTLSSQASDLSWISGRRRQESLGSESEV 0
0 GWTDTEAAAAWGAALQLSGRYLCGQGLEDGEIKATPRRQGPEAETPRK 0
0 TKRLRPCLDPRT* 0
>MEL1_ochPri Ochotona princeps (pika) AAYZ01123854
0 MDPPSGSRVLASPTQEPSSNSSSAAPANRWDSSQNSPSSLGQLSSSSPT 0
0 VLVVQDPFPTVDVPDHAHYTLGSVILLVGLTGMLGNLTVIYTFCR 2
1 SRGLRTPANMFIINLAVSDFLMSFTQAPVFFASSLYKRWIFGET 1
1 GCEFYAFCGALFGISSMITLTAIALDRYLVITRPLAAVGMVSKRRTGLVLLGVWLYSLACSLPPLFGw 1
2 sAYVPEGLLTSCSWDYMTFTPSVRAYTMLLVCFVFFLPLFLIIYCYIFIFRAIRETGR 2
1 VILLFLLSWAPYSTVALVAFAG 2
1 HAHSLTPYMTSVPAVIAKASAIHNPIIYAITHPKYR 2
1 VAIAQHLPCLGVLLGVSGQHSRPSLSYRSTHRSTVSSQTSDLSWISGRRRQESLGSVSEV 0
0 GWADTEVAAAYGASPLVSGRYLYGQVLENEEAKAAPRCHGPGAETPRK 0
0 * 0
>ENC_cavPor Cavia porcellus (guinea pig) 3 aa del
0 MYSGNRSSGQGYWEGGGPEDPAPAGTLSPAPLFSPGAYERLALLLGSLGLLGVGNNLLVLVLYYKFQRLRSPTHLFLANISLSDLLGSLFGVTFTFVSCLKNGWVWDAVGCVWDGFSRSLF 1
2 GIVSITTLTVLAYERYIRVVHARVINFSWAWRAITYIWLYSLAWAGAPLLGWNRYILDIHGLSCTVDWKSKDANDSSFVLFLFLGCLVVPVGVIVHCYGHILYSIRM 0
0 LRGVEDLQTIQVMKILRSENKVAIMCFLMVFIFLVCWMPYIVICFLLVNGYRHRVTPTVSIVSYLFTKSSTVYNPVIYVLMIRK 0
0 FRRSLLQLHCLRLLRCQQPAKDLPAVEREMHIRPIVMSQKDGDRPKKKVTFNSSSIIFIITSDESLSVDDSDRTSGSKVDTIQVRPL* 0
>ENC_musMus Mus musculus (mouse) OPN3 encephalopsin panopsin 2aa del
0 MYSGNRSGDQGYWEDGAGAEGAAPAGTRSPAPLFSPTAYERLALLLGCLALLGVGGNLLVLLLYSKFPRLRTPTHLFLVNLSLGDLLVSLFGVTFTFASCLRNGWVWDAVGCAWDGFSGSLF 1
2 GFVSITTLTVLAYERYIRVVHARVINFSWAWRAITYIWLYSLAWAGAPLLGWNRYILDIHGLGCTVDWRSKDANDSSFVLFLFLGCLVVPVGIIAHCYGHILYSVRM 0
0 LRCVEDLQTIQVIKMLRYEKKVAKMCFLMAFVFLTCWMPYIVTRFLVVNGYGHLVTPTVSIVSYLFAKSSTVYNPVIYIFMNRK 0
0 FRRSLLQLLCFRLLRCQRPAKNLPAAESEMHIRPIVMSQKDGDRPKKKVTFNSSSIIFIITSDESLSVEDSDRSSASKVDVIQVRPL* 0
>ENC_ratNor Rattus norvegicus (rat) XP_573517 2aa del
0 MYSGNRSGGQGYWEDGAGAEGAAPAGTRSPAPLFSPTAYERLALLLGCLALLGVGGNLLVLLLYSKFPRLRTPTHLFLVNLSLGDLLVSLFGVTFTFASCLRNGWVWDAVGCAWDGFSGSLF 1
2 GFVSITTLTVLAYERYIRVVHARVINFSWAWRAITYIWLYSLAWAGAPLLGWNRYILDVHGLGCTVDWKSKDANDSSFVLFLFLGCLVVPMGIIAHCYGHILYSVRM 0
0 LRCVEDLQTIQVIKMLRYEKKVAKMCFLMAFVFLTCWMPYVVTRFLVVNGYGHLVTPTVSIVSYLFAKSSTVYNPVIYIFMIRK 0
0 FRRSLLQLLCFRLLRCQRPAKNLPAAESEMQIRPIVMSQKDGDRPKKKVTFNSSSIIFIITSDESLSVEDSDRSSASKVDVIQVRPL* 0
>ENC_speTri Spermophilus tridecemlineatus (squirrel)
0 MYSGNRSGSQGSWEGDGSAGAEGSAPEGTLSPTPLFSPGTNERLALLFRSVGLLGAGSNLLVLVLYYKFQGSAHPLTFFLVNISLGDLLMSLFGVTFTFVSCLRNRWVWDTVACVWDGFSSSLF 1
2 GIVSITTLTVLAYERYIRVVHARVINFSWAWRAITYIWLYSLAWAGAPLLGWNRYILDVHGLGCTVEWKSKDANDSSFVLFLFLGCLVVPVGVIAHCYGHILYSIRM 0
0 LRCVEDLQIFQVIKILRYEKKLAKMCFVMVFTFLICWMPYIVVCFLVANGYGQRVTPTVSIVSNLFAKSSTVYNPVIYIFMIRK 0
0 FRRSLLQLLCSRLLRCQQPAKDLPAVGNEMQIRPIVISQKDGERPKKKVTFNSSSIVFIITSDESLSVDDSNRTSGSKADVIQVRPL* 0
>ENC_criGri Cricetulus griseus (hamster) XP_003499904=pathetic
0 PTHLFLVNLSLGDLLVSLFGVTFTFASCLRNGWVWDAVGCAWDGFSGSLF 1
2 GFVSITTLTVLAYERYIRVVHSRVINFSWAWRAITYIWLYSLAWAGAPLLGWNRYILDIHGLGCTVDWKSKDANDSSFVLFLFLGCLVVPLGIITHCYGHILYSVRM 0
0 LRCVEDLQTIQVIKILRCEKKVAKMSFAMVFVFLTCWMPYIVTRFLVVNGYGHLVTPTVSIVSSLFAKSSTVYNPIIYIFMIRK 0
0 FRRSLLQLLCFRLLRCQRPAKNLPAAESEMQIRPIVMSQKDRDRPKKKVTFNSSSIIFIITSDESLSVDDSDRTNASKANVIQVRPL
>ENC_dipOrd Dipodomys ordii (kangaroo_rat)
0 MYSGNRSGGQEYWEDGGAAGSEGPAPAGTLSPAPLFSAGAYERLALLLGSAGLLGVGNNLLVLVLYYKFQRLRTPTHLLLVNISLSDLLVSLFGVTFTFVSCLRNGWVWDTVGCVWDGFSRSLF 1
2 GIVSITTLTVLAYERYIRVVHARVINFTWAWRAITYIWLYSLAWAGAPLLGWNRYILDIHGLGCTVDWKAKDANDSSFVLFLFIGCLVVPVGIIAHCYGHILYSIRM 0
0 LRCVEDLQTIQIIKILQYEKKLAKMCFLMALTFLMCWMPYIVTCFLVVNSHGHLVTPTISIVSHLLAKSSTIYNPVIYIFMIRK 0
0 FRRSLLQLLCFRLLRCQRPAKDLPAAGSEMQIRPIVMSQKDGDRPKKKVTFNSSSIIFIITSDESLSVDDSVRSSGSKADVIQVRPL* 0
>ENC_oryCun Oryctolagus cuniculus (rabbit)
0 MYSGNRSGEQGYWEGGGAAGAEGPGPAGTLSPAPLFSPSTYERLALLLGSIGLLGVGSNLLVLVLYYKFQRLRTPTLLFLVNISLSDLLVSVFGVTFTFVSCLRNGWVWDTVGCVWDGFSSSLF 1
2 GIVSITTLTVLAYERYIRVVHARVINFSWAWRAITYIWLYSLAWAGAPLLGWNRYILDIHGLGCTVDWKSKNANDSSFVLFLFLGCLVVPVGVIAHCYGHILYSVRM 0
0 LRCVEDLQTIQVIKILRYEKKVAKMCFFMVFTFLICWMPYVVICFLVVNGYGHLVTPTLSIVSYLFCKSSTAYNPIIYIFMIRK 0
0 FRRSLLQLLCFQPLRCQQPPKDLPTVGSEMQIRPIVMSQKDGDRPKKKVTFNSSSIIFIIASDESLAVDDNEKASGPKVDVIQVRPL* 0
>NEUR1_cavPor Cavia porcellus (guinea_pig)
0 MALNHTAPPQNEHLPRYLQDEDPFVSKLSWEADLVAGFYLTII 1
2 GILSTVGNGYVLYMSSRRKKKLRPAEIMTINLAICDLGIS 1
2 vVGKPFTIISCFRHRWVFGWIGCRWYGWAGFFFGCGSLITMTVVSLDRYLKICYLSY 1
2 GVWLKRKHAYICLAAIWAYVSFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASAGGQIFILHILFFCLLLPTAMIVFSYVKIIAKVKSSSKEIAHFDSRIHSSHVLEMKLTK 0
0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDSRFACCQNAGLKATKKKSLEDFR 2
1 LHTVTTDRKSAVLEIHQEV* 0
>NEUR1_musMus Mus musculus (mouse) OPN5 neuropsin
0 MALNHTALPQDERLPHYLRDEDPFASKLSWEADLVAGFYLTII 1
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1
2 VVGKPFTIISCFCHRWVFGWFGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1
2 GVWLKRKHAYICLAVIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASGGGQVFILSILFFCLLLPTAVIVFSYAKIIAKVKSSSKEVAHFDSRIHSSHVLEVKLTK 0
0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYRFACCQAGGLRGTKKKSLEDFR 2
1 LHTVTTVRKSSAVLEIHQEV* 0
>NEUR1_ratNor Rattus norvegicus (rat)
0 MALNHTALPQDERLPHYLRDEDPFASKLSWEADLVAGFYLTII 1
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1
2 VVGKPFTIISCFCHRWVFGWFGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1
2 gVWLKRKHAYICLAVIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASGGGQVFILSILFFCLLLPTAVIVFSYAKIIAKVKSSSKEVAHFDSRIHSSHVLEVKLTK 0
0 VAMLICAGFLIAWIPYAVVSVWSAFGRPNSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYRFACCQTGGLRATKKKSLEDFR 2
1 LHTVTAVRKSSAVLEIHPEv* 0
>NEUR1_speTri Spermophilus tridecemlineatus (squirrel)
0 MALNHTALPQDEHLPHYLRDEDPFASKLSWEADLVAGFYLTII 1
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1
2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1
2 GVWLKRKHAFICLAVIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASVGGQVFINILFFCLLLPTAVIEFSYVKIIAKVKSSSEEVAHFDSRIHSSHV 0
0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPSLLAKSAAMYNPIIYQVIDYKFACCQTGGLKATKKKSLEDFR 2
1 LHTVTAVRKSSAVVEIHQEv* 0
>NEUR1_dipOrd Dipodomys ordii (kangaroo_rat)
0 MAFNHTAGTQGQGLPHYLPEEDPFTSKLSWEADIVAGFYLTII 1
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1
2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1
2 GVWLKRKHAYICLAVIWAYASFWTTMPLVGLGDYVPEPFGTSCTLDWWLAQASLAGQVFILNILFFCLLLPTSVIVFSYVKIIAKVKSSSKEVAHFDSRIPSSHVLEMKLTK 0
0 2
1 * 0
>NEUR1_oryCun Oryctolagus cuniculus (rabbit)
0 MALNHTALPQDEHLPHYLREGDPFASKLSWEADLVAGFYLTII 1
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1
2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1
2 GVWLKRRHAYICLALIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTK 0
0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFSCCRTSGLKATKKKSLEDFR 2
1 LHTVTTVRKSSAVLEIHQEv* 0
>NEUR1_ochPri Ochotona princeps (pika)
0 MALNDTALPQDEHLPHYFRDGDPFASKLSWEADLVAGFYLTII 1
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1
2 VVGKPFTIISCFCHRWVFGWIGCRLYGWADFFFGCGSLITMTAVSLDRYLK 1
2 GVWLKRRHAYICLAVIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHGSHVLEMKLTK 0
0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFSCCRTGGLKQTKKKSLEDFR 2
1 LHTVTTVRKSSAVLEIHQEv* 0
>PER_cavPor Cavia porcellus (guinea_pig)
0 MLRHSLGNSSDSKNEDGSVFSQTEHNIVAAYLILA 1
2 GLISILSNIIVLGIFIKYKELRTPTNAIIMNLALTDIGVSSIGYPMSAASDLHGSWKFGYAGCQ 0
0 VYAGLNIFFGMASIGLLTVVAVDRYLTICRPDI 1
2 GRRMTSHSYVGMILGAWINGLFWALMPIIGWASYAPDPTGATCTINWRKNDV 2
1 AFVSYTMTVIAINFIVPLAVMFYCYLHITRAIRRHVAGDRPPNLSGDWSDQVDVTK 0
0 MSVVMILMFLVAWSPYSIVCLWASFGDPRRISPSMAIIAPLFAKSSTFYNPCIYVIANKK 2
1 FRRAMFAMFQCQTHQAVPVASILPMDASQSPLASGRI* 0
>PER_musMus Mus musculus (mouse) RRH peropsin syn(-CFI +NOLA1 +EGF -ELOVL6)
0 MLSEASDNSSGSRSEGSVFSRTEHSVIAAYLIVA 1
2 GITSILSNVVVLGIFIKYKELRTPTNAVIINLAFTDIGVSSIGYPMSAASDLHGSWKFGHAGCQ 0
0 IYAGLNIFFGMVSIGLLTVVAMDRYLTISCPDV 1
2 GRRMTTNTYLSMILGAWINGLFWALMPIIGWASYAPDPTGATCTINWRNNDT 2
1 SFVSYTMMVIVVNFIVPLTVMFYCYYHVSRSLRLYAASDCTAHLHRDWADQADVTK 0
0 MSVIMILMFLLAWSPYSIVCLWACFGNPKKIPPSMAIIAPLFAKSSTFYNPCIYVAAHKK 2
1 FRKAMLAMFKCQPHLAVPEPSTLPMDMPQSSLAPVRI* 0
>PER_ratNor Rattus norvegicus (rat)
0 MLRDALDNSSGSGSE-GSVFTKSEHSIIAAYLIVA 1
2 GIISILSNIIVLGIFIKYKELRTPTNAVIINLAFTDIGVSSIGYPMSAASDLHGSWKFGHAGCQ 0
0 VYAGLNIFFGMVSIGLLTVVALDRYLTISCPDV 1
2 GRRMTGNTYLSMVLGAWINGLFWALMPIVGWASYAPDPTGATCTINWRKNDT 2
1 SFVSYTMMVIVVNFIVPLTVMFYCYYHVSQSMRLSAASNCTTHLNRDWAHQADVTK 0
0 MSVMMILMFLLAWSPYSVVCLWACFGNPKKIPPSLAIIAPLFAKSSTFYNPCIYVAANKK 2
1 FRKAMFAMLKCQPHQAMPEPSTLAMGVPHSPLAPARI* 0
>PER_dipOrd Dipodomys ordii (kangaroo_rat)
0 MLRNNVGNSSGSRNEDGSVFSQTEHNIVATYLITA 1
2 GVISILSNLIVLGIFIKYKELRTPTNAIIINLALTDIGVSSIGYPMSAASDLYGRWKFGYAGCQ 0
0 IYAGLNIFFGMASIGLLTVVAIDRYLTICHPDI 1
2 GRGMTTRTYVTMILGAWINGLFWALMPIIGWASYAPDPTGATCTINWRKNDT 2
1 0
0 MSVVMILMFLVAWSPYSIVCLWASFGDPKEIPPPMAIIAPLFAKSSTFYNPCIYVVANKK 2
1 FRRAMLAMLKCQTHQAMPVTSILPMDVSQNPLASGRI* 0
>PER_oryCun Oryctolagus cuniculus (rabbit)
0 MLRNNLSNSSDFKHEDGSVFSQTEHNIVATYLILA 1
2 GMISILSNLIVLGIFIKYKELRTPTNAIIINLAFTDIGVSSIGYPMSAASDLHGSWKFGYAGCQ 0
0 IYAGLNIFFGMASIGLLTVVAMDRYLTICHPDV 1
2 GRRMTTRTYLGLILGAWVNGLFWALMPIAGWASYAPDPTGATCTINWRKNDT 2
1 SFVSFTMAVIAINFVVPLTVMFYCYYHVTQSIKQHRASDCTEYLNRDWSDQVDVTK 0
0 MSVIMIFMFLVAWSPYSIVCLWASFGDPKKIPPAMAIIAPLFAKSSTFYNPCIYVAANKR 2
1 FRRAMFAMFKCQTHQAMPVTSVLPMDVSQNPLPSGII* 0
>PER_ochPri Ochotona princeps (pika)
0 MLRHNLGNSSEAKVEAGSVFSQTEHNIVAAYLILA 1
2 GMISILSNLIVLGIFIKYKELRTPTNAIIINLAFTDIGVSSIGYPMSAASDLHGSWKFGYAGCQ 0
0 IYAGLNIFFGMASIGLLTVVAMDRYLTICQPDI 1
2 GRRMTTHTYFGMILGAWINGLFWALMPIVGWASYAPDPTGATCTINWRKNDK 2
1 SFVSFTMAVIMVNFVVPLTVMFYCYYYVTQSIKHHTASDCTKSLNRDWSDQVDVTK 0
0 MSVIMILMFLVAWSPYSIVCLWASFGDPQKIPPSMAIIAPLFAKSSTFYNPCIYVAANKR 2
1 SRRAMFAMFKCQIPQAKPVTSLSPRDVSQSPLSSGRT* 0
>RGR_cavPor Cavia porcellus (guinea_pig)
0 MATSEALPAGFGELEVLAVGTVLLLE 1
2 GLCGLSLNGLTVVSFWKSPALRTPNHLLVLSLALADSGLSLNALVAAGSSLLR 2
1 HWPGSGHCQALGFQGFTTALASISGTAALSWGRHQQCCT 1
2 RGRLTWSTAVPLVLFVWLSSAFWAALPLLGWGRYDYEPLGTCCTLDYSTGDR 2
1 NFTSFLFTMAFFNFLVPLFITVTSCQLMERHLARSSRLQ 0
0 VSVRQPARTLLLCWSPYALLYLYAVLADAHTLSPRLQM 0
0 VPALIAKTVPTIYSLGRGPWQSLEMQRSKQD* 0
>RGR_musMus Mus musculus (mouse) RGR syn(+PCDH21 -LRIT1 -GRID1 -WAPAL)
0 MAATRALPAGLGELEVLAVGTVLLME 1
2 ALSGISLNGLTIFSFCKTPDLRTPSNLLVLSLALADTGISLNALVAAVSSLLR 2
1 RWPHGSEGCQVHGFQGFATALASICGSAAVAWGRYHHYCT 1
2 GRQLAWDTAIPLVLFVWMSSAFWASLPLMGWGHYDYEPVGTCCTLDYSRGDR 2
1 NFISFLFTMAFFNFLVPLFITHTSYRFMEQKFSRSGHLP 0
0 VNTTLPGRMLLLGWGPYALLYLYAAIADVSFISPKLQM 0
0 VPALIAKTMPTINAINYALHREMVCRGTWQCLSPQKSKKDRTQ* 0
>RGR_ratNor Rattus norvegicus (rat)
0 MTATRALPAGFGELEVLAIGIVLLME 1
2 ALSGISLNGLTIFSFCKTPDLRTPSNLLVLSLALADTGISLNALVAAVSSLLR 2
1 RWPHGSEGCRVHGFQGFATALASICGSAAIAWGRYHHYCT 1
2 GRQLAWDTAIPLVLFVWLSSAFWASLPLMGWGHYDYEPVGTCCTLDYSRGDR 2
1 NFISFLFTMAFFNFLVPLFITHTSYRFMEQKLSRSGHLQ 0
0 VNTTLPGRMLLLGWGPYALLYLYAAVADVSFISPKLQM 0
0 VPALIAKTMPTINAINYALRSEMVCRGTWQCRSAQKSKQDRTQ* 0
>RGR_speTri Spermophilus tridecemlineatus (ground_squirrel)
0 MAETAALPAGFGELEVLAVGTVLLVE 1
2 ALSGLSLNGLTIFSFCKTPELRTPNHLLLLSLAVADSGISLNALIAAISSLLR 2
1 RWPYGSDGCQAHGFQGFVTALSSICGSAAIAWGRYHHYCT 1
2 GSQLAWNTAIPLVLFVWLSSTFWAALPLLGWGHYDYEPLGTCCTLDYSRGDR 2
1 NFISFLFTMAFFNFFVPLFITLTSYRLMEQKLARSGHLQ 0
0 0
0 * 0
>RGR_dipOrd Dipodomys ordii (kangaroo_rat)
0 MATSGDLPTGFGELEVLTVGTVLLVE 1
2 ALSGLSLNTLTIFSFCKTPELRTPIHLLDLSLAVADSGISLNALIAAISSLEW 2
1 HWPYGLEGCQAHGFQGFVTALASISGSAAIAWGRCHHHCT 1
2 GSLLGWDTAVSLVIFVWLSSAFWAALPLLGWGHYNYEPLGTCCTLDYSRGDR 2
1 NFTSFLFTMAFFNFLVPLFITLTSYQLMKQKFARSGRLQ 0
0 VNTTLPTRTLLLGWGPYALLYFYAAIMDVNSISPKLQM 0
0 VPALIAKMVPTVNAINYALCNELLCGGFSLGLLPQKGKQDRTQ* 0
>RGR_oryCun Oryctolagus cuniculus (rabbit)
0 MAEPGTLPPGFEELEVLAVGTVLLVE 1
2 ALSGLSLNGLTIFSFCKTPELWTPSHLLVLSLAVADSGISLNALIAAVSSLLR 2
1 RWPYGSDGCQAHGFQGFATALASICGSAAIAWGRYHHYCT 1
2 GSQLAWNTAVLLVLFVWLSSVFWAALPLLGWGHYDYEPLGTCCTLDYSRGDR 2
1 NFISFLITMAFFNFLMPLFITLTSYSLMEQKLSKSGRLQ 0
0 VNTTLPGRTLLFCWGPYAVLYLCAAVADMSSITLKLQM 0
0 VPALIAKTVPTVNAVNYALGSEVIRRGIWQCLLPQRSVRGRAQ* 0
>RGR_ochPri Ochotona princeps (pika)
0 MAEPGTLPPGFEELEVLAVGTVLLVE 1
2 ALTGLSLNSLTIFSFCTSPELRTPSHLLVLSLALADSGVSLNALAAATASLLR 2
1 RWPYGSDGCQAHGFQGFATALASICGSAAIAWGRYHHYCT 1
2 GSQLAWNTAVLLVLFVWLSSVFWAALPLLGWGHYDYEPLGTCCTLDYSRGDR 2
1 NFISFLVTMAFFNFLMPLFIMLTSYSLMEQKLAKSGRLQ 0
0 VNTTLPARTLLFCWGPYAILCLCATVMDMSTVSPKLLM 0
0 VPALIAKAVPTVNAINYALGSEVIRRGIWQCLLPQRSVRDRAQ* 0
Rodent cryptochromes
>CRY2_hetGla Heterocephalus glaber (mole-rat) EHA99865 MAAAVGTGTGAAPTPATGAEGACSVHWFRKGLRLHDNPALLAAVRGARCVRCVYILDPWFAASSSVGINRW RFLLQSLEDLDTSLRKLNSRLFVVRGQPADVFPRLFK EWGVTRLTFEYDSEPFGKERDAAIMKMAKEAGVEVVTENSHTLYDLDR IIELNGQKPPLTYKRFQAIISRMELPKKPVGAVSSQQMKSCRAEIQENHDDTYGVPSLEEL GFPTEGLGPAVWQGGETEALARLDKHLERK AWVANYERPRMNANSLLASPTGLSPYLRFGCLSCRLFYYRLWDLYKK VKRNSTPPLSLFGQLLWREFFYTAATNNPRFDRMEGNPICIQIPWDRNPEALAKWAEGKTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWESGVR VFDELLLDADFSVNAGSWMWLSCSAFFQQFFHCYCPVGFGRRTDPSGDYIR RYLPKLKGFPSRYIYEPWNAPESVQKAAKCIIGVDYPRPIVNHAETSRLNIERMKQIYQQLSRYRGL CLLASVPSCVEDLSHPVAEPTLSQAGSSSSTG PRSLPDGPASPKRKLEAAEEPPGEEL SKRARVAELPAPEPTSKDA* >CRY1_hetGla Heterocephalus glaber (mole-rat) no workable iMet, stop codon at position 8 in place of conserved W, last two exons very diverged 0 MAMNAVH*FRKGLRLHDNPALKECTRGADTIRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNITKLSIEYDSEPFGKERDAAIKKLATEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLISKMEPLEIPVETITSEVIEKCVTPLSDDHDEKYGVPSLEEL 1 2 GFDTDGLPSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNGNGGLMGYAPGESIPGSSGSG 1 2 SCAHGSGILPCAHTDGQQAHLLKP 1 2 GRNCVGPVLSSGKRPSQEEDAQSIGPKLQRQSTD* 0 >CRY1_cavPor Cavia porcellus (guinea pig) last two exons diverged 690 bp separation agttgcgtccccgggagcagcagcgcgggagtctcgcactgtgcgcagggagaagctccgcaggcgccgccag 0 MGVNAVHWFRKGLRLHDNPALKECIRGADTIRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNITKLSIEYDSEPFGKERDAAIKKLASEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLISKMEPLEIPVETITSEVVEKCVTPLSDDHDEKYGVPSLEEL 1 2 GFDTDGLPSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDRNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWVSWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVHHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNGNGGLLGYAPGESTPGSGGG 1 2 SCVPGSSSAGVSHCAQGEAPQAPP 1 2 GRDPAGPGLGGGKRPSQEEDAQSTGHKIQRQSPD* 0 >CRY1_musMus Mus musculus (mouse) NM_007771 all transcripts support longer exon 10 lost splice donor 0 MGVNAVHWFRKGLRLHDNPALKECIQGADTIRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNITKLSIEYDSEPFGKERDAAIKKLATEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLVSKMEPLEMPADTITSDVIGKCMTPLSDDHDEKYGVPSLEEL 1 2 GFDTDGLSSAVWPGGETEALTRLERHLER 0 0 KAWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNSNGNGGLMGYAPGENVPSCSSSGNGGLMGYAPGENVPSCSGG 1 2 NCSQGSGILHYAHGDSQQTHSLKQ 1 2 GRSSAGTGLSSGKRPSQEEDAQSVGPKVQRQSSN* 0 >CRY1_ratNor Rattus norvegicus (rat) NM_198750 0 MGVNAVHWFRKGLRLHDNPALKECIQGADTIRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNITKLSIEYDSEPFGKERDAAIKKLATEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLVSKMEPLEMPADTITSDVIGKCTTPLSDDHDEKYGVPSLEEL 1 2 GFDTDGLSSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNGNGGLMGYAPGENVPSGGSGGG 1 2 NCSQGSGILHYAHGDSQQTNPLKQ 1 2 GRSSMGTGLSSGKRPSQEEDAQSVGPKVQRQSSN* 0 >CRY1_criGri Cricetulus griseus (hamster) XM_003505292 0 MGVNAVHWFRKglrlhDNPALKECIQGADTIRCVYILDPWFAGSSNVGINRWR 2 1 FLLQCLEDLDSNLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNITKLSIEYDSEPFGKERDAAIKKLATEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLVSKMEPLEMPAETITSDVIGKCMTPLSDDHDEKYGVPSLEEL 1 2 GFDTDGLSSAVWPGGETEALTRLERHLER 0 0 KAWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNGNGGLMGYTTGENLPSCSGGG 1 2 SCSQGSGILHYAHGDSQQAHLLKQ 1 2 GRSSMGTSLSSGKRPSQEEETRSVDPKVQRQSSN* 0 >CRY1_spaJud Spalax judaei (blind mole rat) AJ606298 0 MGVNAVHWFRKGLRLHDNPALKECIQGADTIRCVYILDPWFAGSSNVGINRW 2 1 RFLLQCLEDLDANLRKLNSRLLVIRGQPADVFPRLFKEWNITKLSIEYDSEPFGKERDAAIKKLATEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLISKMEPLEIPVETITSEVIEKCTTPLSDDHDEKYGVPSLEEL 1 2 GFDTDGLSSAVWPGGETEALTRLERHLERR 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTGSDLWISWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNGNGGLMGYTPGENIPNCSSSG 1 2 SCSQGSGILHYAHGDSQQAHLLKQ 1 2 GSSSMGHGLSNGKRPSQEEDTQSIGPKVQRQSTN* 0 >CRY1_dipOrd Dipodomys ordii (kangaroo rat) ABRO01202522 ABRO01202521 0 MGVNAVHWFRKGLRLHDNPALKECIQGADTIRCVYILDPWFAGSSNVGINRW 2 1 RFLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNITKLSIEYDSEPFGKERDAAIKKLATEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLISKMEPLEIPVETITSEVIEKCTTPLSDDHDEKYGVPSLEEL 1 2 GFDTDGLSSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCSVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNGNGGLMGYAAGDNLPGSSSSG 1 2 SCSQGSGILHYAHGDSQQMHLLKQ 1 2 GRSSMGTGLSSGKRPSQEEDSQSIGPKVQRQSTN* 0 >CRY1_speTri Spermophilus tridecemlineatus (squirrel) Ictidomys 0 2 1 0 RDAAIKKLATEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLISKMEPLEIPVETITSEVIEKCTTPLSDDHDEKYGVPSLEEL 1 2 GFDTDGLSSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGVNYPKPMVNHEASLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNGNGGLMAyAPGENIPGCSSSG 1 2 SCTQGSSILHNAHGDSQQTHLLKQ 1 2 GRSSMGTGLSSGKRPSQEEDTQSIGPKVQRQSTN* 0 >CRY1_oryCun Oryctolagus cuniculus (rabbit) 0 MGVNAVHWFRKGLRLHDNPALKECIQGADTIRCVYILDPWFAGSSNVGINRW 2 1 RFLLQCLEDLDANLRKLNSRLFVIRGQPADVFPRLFK 0 0 EWNITRLSIEYDSEPFGKERDAAIKKLASEAGVEVIVRISHTLYDLDK 2 1 IIELNGGQPPLTYKRFQTLISKMEPLEIPVETITSDVIEKCTTPVSDDHDEKYGVPSLEEL 1 2 GFDTDGLSSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGQTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 VFEELLLDADWSINAGSWMWLSCSSFFQQFFHCYCPVGFGRRTDPNGDYIR 2 1 RYLPVLRGFPAKYIYDPWNAPEGIQKVAKCLIGINYPKPMVNHAEASRLNIERMKQIYQQLSRYRGL 1 2 GLLASVPSNPNGNGGLMGYSPGENIPGCSSSG 1 2 SCSQGSGILHYAQGDTQQTQLLKQ 1 2 GRSSMGTGLSSGKRPSQEEDTQSIGPKVQRQSTN* 0 >CRY1_ochPri Ochotona princeps (pika) 0 2 1 0 0 EWNITKLSIEYDSEPFGKERDAAIKKLATEAGVEVIVRISHTLYDLDK 2 1 1 2 GFDTDGLSSAVWPGGETEALTRLERHLERK 0 0 AWVANFERPRMNANSLLASPTGLSPYLRFGCLSCRLFYFKLTDLYKK 0 0 VKKNSSPPLSLYGQLLWREFFYTAATNNPRFDKMEGNPICVQIPWDKNPEALAKWAEGRTGFPWIDAIMTQLRQEGWIHHLARHAVACFLTRGDLWISWEEGMK 0 0 2 1 1 2 1 2 GCPQGSGILHCGHGDSQQTHLLKQ 1 2 GRGSPDTGISSGKRPSQEEDTQSIGPKVQRQSTN* 0