Opsins underground: Difference between revisions
Tomemerald (talk | contribs) |
Tomemerald (talk | contribs) |
||
| Line 17: | Line 17: | ||
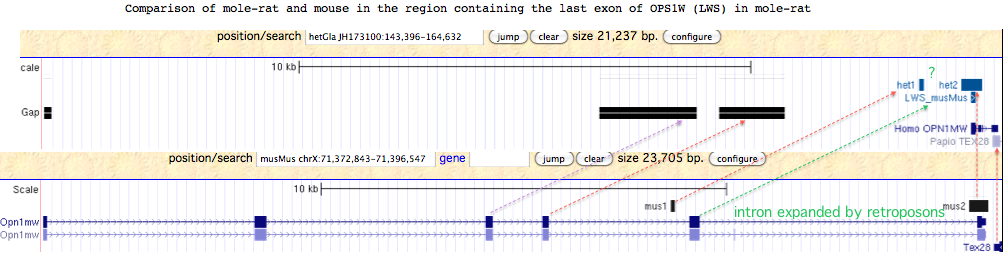
The second line is also in error. The terminal exon of mole-rat OPN1LW is readily recovered by Blat of mouse query against mole-rat assembly. This gene fragment occurs in mole-rat in the long-conserved syntenic location IRAK1 MECP2 OPN1LW TEX28. The amino acid sequence is quite conserved and not suggestive of pseudogenization. Since it lies in a region with several large assembly gaps corresponding to the missing exon placement in mouse genome, it follows that mole-rat very likely has a complete functional copy of OPN1LW. However it is not clear why the penultimate exon 5 cannot be recovered by blastx from the region indicated by the green arrow. | The second line is also in error. The terminal exon of mole-rat OPN1LW is readily recovered by Blat of mouse query against mole-rat assembly. This gene fragment occurs in mole-rat in the long-conserved syntenic location IRAK1 MECP2 OPN1LW TEX28. The amino acid sequence is quite conserved and not suggestive of pseudogenization. Since it lies in a region with several large assembly gaps corresponding to the missing exon placement in mouse genome, it follows that mole-rat very likely has a complete functional copy of OPN1LW. However it is not clear why the penultimate exon 5 cannot be recovered by blastx from the region indicated by the green arrow. | ||
OPN1LW LWS_hetGla FPNCILQLFGKKVEDSLELSSASKTEASSVSSVSPA* last exon aligned to mouse | |||
F.NCIL.LFGKKV.DS.ELSS.SKTE.SSVSSVSPA* | |||
OPN1LW LWS_musMus FRNCILHLFGKKVDDSSELSSTSKTEVSSVSSVSPA* | |||
[[Image:MoleRATlws.png]] | [[Image:MoleRATlws.png]] | ||
| Line 31: | Line 31: | ||
Gene Protein Description Inactivation event Time of gene loss | Gene Protein Description Inactivation event Time of gene loss | ||
<font color = red>OPN1MW MWS mid wavelength complete loss detected with synteny within NMR stem lineage | <font color = red>OPN1MW MWS mid wavelength complete loss detected with synteny within NMR stem lineage | ||
OPN1LW LWS long wavelength | OPN1LW LWS long wavelength complete loss detected with synteny within NMR stem lineage</font> | ||
OPN1SW SWS1 short wavelength none retained to present | |||
RHO RHO1 dim rhodopsin none retained to present | RHO RHO1 dim rhodopsin none retained to present | ||
OPN3 ENC encephalopsin none retained to present | OPN3 ENC encephalopsin none retained to present | ||
OPN4 MEL1 melanopsin none retained to present | OPN4 MEL1 melanopsin none retained to present | ||
RRH PER peropsin none retained to present | RRH PER peropsin none retained to present | ||
OPN5 NEUR1 neuropsin none retained to present | OPN5 NEUR1 neuropsin none retained to present | ||
Revision as of 17:32, 15 February 2012
Introduction
The genome article for the naked mole-rat (Heterocephalus glaber) appeared in Nature in October, 2011. That paper made some remarkable observations about aging in this species, the longest-lived of all rodents.
Because the mole-rat has a very unusual life history, spending its entire life underground, the question arises as to what has happened to the eight phylogenetically expected opsins. The Nature article addresses this, but only to an extent constrained by the many other genes needing commentary.
According to the article,
We further examined the molecular basis for poor visual function and small eyes in the NMR (naked mole-rat). Of the four vertebrate opsin genes (RHO, OPN1LW, OPN1MW and OPN1SW), two (OPN1LW and OPN1MW) were missing; this distinguishes the NMR from other rodents with dichromatic colour vision, such as mice, rats and guinea pigs. However, the NMR has intact RHO (rhodopsin) and OPN4 (melanopsin), supporting the presence of rod-dominated retinae and the capacity to distinguish light/dark cues.
Of about 200 genes associated with visual perception (GO:0007601) in humans and mice, almost 10% were inactivated or missing. These mammalian genes participate in crystallin formation, phototransduction in the retina, retinal development, dark adaptation, night blindness and colour vision.... Inactivation of CRYBA4, a microphthalmia-related gene, may be associated with the small-sized eyes, whereas inactivation of CRYBA4 and CRYBB3 and a NMR-specific mutation in CRYGS may be associated with abnormal eye morphology. Thus, while some genes responsible for vision are preserved, poor visual function may be explained by deterioration of genes coding for various critical components of the visual system.
The first two lines of the summary table are reproduced from the article. The others arise from consideration of mammalian opsin repertoires, in particular those of mouse and orthologous counterparts found by methods described here. Note the first line OPN1MW is an outright error as this gene is well-known to be a tandem duplication restricted to higher primates with no applicability to other mammals. It never existed in any ancestor of mole-rat and so was never lost.
The second line is also in error. The terminal exon of mole-rat OPN1LW is readily recovered by Blat of mouse query against mole-rat assembly. This gene fragment occurs in mole-rat in the long-conserved syntenic location IRAK1 MECP2 OPN1LW TEX28. The amino acid sequence is quite conserved and not suggestive of pseudogenization. Since it lies in a region with several large assembly gaps corresponding to the missing exon placement in mouse genome, it follows that mole-rat very likely has a complete functional copy of OPN1LW. However it is not clear why the penultimate exon 5 cannot be recovered by blastx from the region indicated by the green arrow.
OPN1LW LWS_hetGla FPNCILQLFGKKVEDSLELSSASKTEASSVSSVSPA* last exon aligned to mouse
F.NCIL.LFGKKV.DS.ELSS.SKTE.SSVSSVSPA*
OPN1LW LWS_musMus FRNCILHLFGKKVDDSSELSSTSKTEVSSVSSVSPA*
The article did not consider OPN1SW. This cone opsin is present and little diverged from available rodent orthologs suggesting functionality, with the exception of a six residue deletion in exon 4 associated with a frameshift. This occurs following a run of eight thymidines TTTTTTTT and probably represents inability of short read technology to precede past a compositionally simple region and allow assembly, rather than an actual deletion.
Thus like most placental mammals, mole-rat has the genetic potential for two-color cone vision plus dim-light rhodopsin, in addition to other opsins whose anatomical sites of expression (in the absence of mole-rat data) are likely the same as in mouse. The functions of these other opsins -- with the exception of rhabdomeric melanopsin -- are poorly understood.
This article henceforth will use protein acronyms because assigned gene names have no mneumonic value and do not follow HGNC rules for paralog nomenclature.
Gene Protein Description Inactivation event Time of gene loss OPN1MW MWS mid wavelength complete loss detected with synteny within NMR stem lineage OPN1LW LWS long wavelength complete loss detected with synteny within NMR stem lineage OPN1SW SWS1 short wavelength none retained to present RHO RHO1 dim rhodopsin none retained to present OPN3 ENC encephalopsin none retained to present OPN4 MEL1 melanopsin none retained to present RRH PER peropsin none retained to present OPN5 NEUR1 neuropsin none retained to present RGR RGR rgropsin none retained to present
Rodent opsin reference sequences
Like all genome assemblies, that of molerat has incomplete coverage. Here eight genes were expected and using mouse probes -- eight were detected. However only 11 of the 50 exons expected were represented (78% coverage). The comparative genomics of opsins have been studied to exhaustion: no variation in exon number or intron location or phasing occurs in any terrestrial vertebrate.)
The sequences below have one exon per row. The numbers 0 1 2 indicate the observed codon overhang (reading frame). These are deeply conserved within eumetazoans; mole-rat presents no new issues. Processed pseudogenes are absent, consistent with no gene expression of opsins in terminal germline cell lineages.
>ENC_hetGla Heterocephalus glaber (naked molerat) OPN3 88% identical musMus, missing first exon 0 1 2 GIVSITTLTVLAYERYIRVVHARVINFSWAWKAITYIWLYSLAWAGAPLLGWNRYILDVHGLSCTVDWKSKDANDSSFVLFLFLGCLVVPMGVIAHCYGHILYSIRM 0 0 FSMLQLRCVEDLQTIQVMKILRYEKKVAKMCFLMVFIFLVCWMPYIVICFLVVNGYGHRVTPTVSVVSYLFAKSSTVYNPVIYTIMIRK 0 0 FRRSLLQLLCFRLLRCQQPAKDLPAVESEMQIRPIVMSQKDGDRPKKKVTFNSSSIIFIITSDESLSVDDSDRTSGSKVDTIQVRPL* 0 >LWS_hetGla Heterocephalus glaber (naked molerat) OPN1LW syn(-IRAK1 -MECP2 -TEX28) 88% identical musMus, missing exons 1-5 because of assembly gaps 0 1 2 1 2 2 1 2 0 0 0 FPNCILQLFGKKVEDSLELSSASKTEASSVSSVSPA* 0 >SWS1_hetGla Heterocephalus glaber (naked molerat) OPN1SW syn(-FAM137A +SWS1 -CALU -NAG6 -FLNC) 89% identical musMus, frameshifting deletion 6aa exon 4 0 MSGEEDFYLFKNTSSVGPWDGSQYHIAPIWAFHLQAAFMGLVFFVGTPLNAIVLVATLQYKKLRQPLNYILVNVSLGGFLFCIFSVFTVFIASCHGYFFFGRHVCALEAFLGSVA 1 2 GLVTGWSLAFLAFERYLVICKPFGNFRFSSKHALIVVLATWVIGIGVSIPPFFGWSR 2 1 YIPEGLQCSCGPDWYTVGTKYRSEYYSWFLFIFCFIVPLSLICFSYSQLLRTLRA 0 0 VAAQQQESATTQKAEREVSRMVVVMVGSFCVCYVPYAALAMYIVNNRNHGLDLRLFTIPAFFSPKSCVYNPIIYCFMNKQ 0 0 FRACIMELVCRKSMADESDMSSSQKTEVSAVSSSKVGPN* 0 >RHO1_hetGla Heterocephalus glaber (naked molerat) RHO syn(-MBD4 +IFT122 +H1FOO -PLXND1) MNGTEGPNFYVPFSNVTGVVRSPFEQPQYYLAEPWQFSMLAAYMFLLIVLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLG GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVVFTWIMALACAAPPLVGWSR YIPEGMQCSCGIDYYTLKPEVNNESFVIYMFVVHFTIPMIVIFFCYGQLVFTVKE AAAQQQESATTQKAEKEVTRMVIIMVIFFLICWLPYASVAFYIFTHQGSNFGPIFMTLPAFFAKSSSIYNPVIYIMLNKQ FRNCMLTTLCCGKNPLGDDDASATASKTETSQVAPA >MEL1_hetGla Heterocephalus glaber (naked molerat) OPN4 86% identical musMus, missing exons 1,8,9 0 0 0 AAGTQAAVRVPFSTADVPAHVHYTLGAVILLVGLTGMLGNLTVIYTFFR 2 1 RGLRTPANMFVINLAVSDFLMSFTQAPVFFTSSLYKRWLFGEA 2 2 GCEFYAFCGALFGITSMITLTAITLDRYLVITRPLATIGLASKRQAALVLLGIWLYALAWSLPPFFGW 1 2 sAYVPEGLLTSCSWDYMTFTPSVRSYTMLLFCFVFFLPLLIIIYCYVFIFRAIRETGR 2 1 AFESCGESPSRRRQWQRLRSEWKMAKIALLVILLFVLSWAPYSTVALVAFAG 2 1 HAHILTPSMSSVPAVIAKASAIHNPIIYAIAHPKYR 2 1 MAIVQHLPCLGLLLGVSGQRSLPSLSYRSTHRSRLSSQASDLSWISGRRRQESLGSESEV 0 0 0 0 * 0 >PER_hetGla Heterocephalus glaber (naked molerat) RRH syn(-CFI +NOLA1 +EGF -ELOVL6) 79% identical musMus, missing exons 1,7 0 1 2 GLISILSNIIVLGIFVKYKELRTPTNAIIMNLALTDIGVSSIGYPMSAASYLHGSWKFGYPGCQ 0 0 VYAGLNIFFGMASIGLLTVIAVDRYLTICRPDI 1 2 GRRLTSTSYVSMILGAWINGLFWALMPIIGWASYAPDPTGATCTINWRKNDt 2 1 SFVSYTMTVIAVNFTGPLAVMFYCYFHVTWSIKHHATGNCPTFPNRDWSDQVDVTK 0 0 MSVVMILMFLVAWSPYSIVCLWASFGDPKKIPPSMAIIAPLFAKSSTFYNPCIYVIANKK 2 1 * 0 >NEUR1_hetGla Heterocephalus glaber (naked molerat) OPN5 94% identical musMus 0 MALNHTAPPQDERLPHYLQDEDPFVSKLSWEADLVAGFYLTII 1 2 GILSTCGNGYVLYMSSRRKKKLRPAEIMTINLAICDLGIS 1 2 VVGKPFTIISCFRHRWVFGWIGCRWYGWAGFFFGCGSLTTMTVVSLDRYLKICYLSY 1 2 GVWLKRKHAYICLAAIWAYVSFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASARGQVFILNILFFCLLLPMATIVFSYAKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYRFACCQNGGLKATKKKSLEDFR 2 1 LHTVTTVRKSSAVLEIHQEV* 0 >RGR_hetGla Heterocephalus glaber (naked molerat) RGR syn(+PCDH21 -LRIT1 -GRID1 -WAPAL) 74% identical musMus, missing exons 1 0 1 2 ALCGLSLNGLTIVSFCKIPELWTPRHLLVLSLALADSGLSLNTLIAAVSSLLR 2 1 RWPYGSGGCQTHGFQGFVTALASISGSAAIAWGHHQHYCT 1 2 GSQLAWSTAIRLVLFVWLSSAFWAALPLLGWGHYDYEPLGTCCTLDYSKRDR 2 1 NSTSFLFTMGFFSFLIPLFITFTSYQLMEQKLARSSRLQ 0 0 VNTSLPARTLMLCWGPYVFLHLYAVIADASSLSPKLQM 0 0 VPALIAKTVPTINAINYALGSKMGSRRPWQCLSLQRSKRD* 0 >ENC_musMus Mus musculus (mouse) OPN3 encephalopsin panopsin 2aa del 0 MYSGNRSGDQGYWEDGAGAEGAAPAGTRSPAPLFSPTAYERLALLLGCLALLGVGGNLLVLLLYSKFPRLRTPTHLFLVNLSLGDLLVSLFGVTFTFASCLRNGWVWDAVGCAWDGFSGSLF 1 2 GFVSITTLTVLAYERYIRVVHARVINFSWAWRAITYIWLYSLAWAGAPLLGWNRYILDIHGLGCTVDWRSKDANDSSFVLFLFLGCLVVPVGIIAHCYGHILYSVRM 0 0 LRCVEDLQTIQVIKMLRYEKKVAKMCFLMAFVFLTCWMPYIVTRFLVVNGYGHLVTPTVSIVSYLFAKSSTVYNPVIYIFMNRK 0 0 FRRSLLQLLCFRLLRCQRPAKNLPAAESEMHIRPIVMSQKDGDRPKKKVTFNSSSIIFIITSDESLSVEDSDRSSASKVDVIQVRPL* 0 >LWS_nanEhr Nannospalax ehrenbergi (mole_rat) OPN1LW syn(-IRAK1 -MECP2 -TEX28) 0 MAQQWAPQRLAGGQTQDSYEDSTQASLFTYTNSNSTR 1 2 GPFEGPNYHIAPRWVYHLTTTWMILVVVASIFTNGLVLVATMRFKKLRHPLNWILVNLAVADLAETIIASTISVVNQIYGYFVLGHPLCVIEGYTVSLC 1 2 GITGLWSLAIISWERWMVVCKPFGNVRFDAKLATMGITFAWVWAAVWTAPPVFGWSR 2 1 YWPYGLKTSCGPDVFSGTSYPGVQSYMVVLMITCCIIPLSIILLCYLQVWLAIRT 2 0 VAKQQKESESTQKAEKEVTHMVVVMVFAYCLCWGPYTFFVCFATAHPGYAFHPLVAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FQNCILQLFGKKVDDSSELSSTSKTEASSVSSVSPA* 0 >SWS1_musMus Mus musculus (mouse) OPN1SW syn(-FAM137A +SWS1 -CALU -NAG6 -FLNC) 0 MSGEDDFYLFQNISSVGPWDGPQYHLAPVWAFRLQAAFMGFVFFVGTPLNAIVLVATLHYKKLRQPLNYILVNVSLGGFLFCIFSVFTVFIASCHGYFLFGRHVCALEAFLGSVA 1 2 GLVTGWSLAFLAFERYVVICKPFGSIRFNSKHALMVVLATWIIGIGVSIPPFFGWSR 2 1 FIPEGLQCSCGPDWYTVGTKYRSEYYTWFLFIFCFIIPLSLICFSYSQLLRTLRA 0 0 VAAQQQESATTQKAEREVSHMVVVMVGSFCLCYVPYAALAMYMVNNRNHGLDLRLVTIPAFFSKSSCVYNPIIYCFMNKQ 0 0 FRACILEMVCRKPMADESDVSGSQKTEVSTVSSSKVGPH* 0 >RHO1_musMus Mus musculus (mouse) RHO syn(-MBD4 +IFT122 +H1FOO -PLXND1) 0 MNGTEGPNFYVPFSNVTGVVRSPFEQPQYYLAEPWQFSMLAAYMFLLIVLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLG 1 2 GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVVFTWIMALACAAPPLVGWSR 2 1 YIPEGMQCSCGIDYYTLKPEVNNESFVIYMFVVHFTIPMIVIFFCYGQLVFTVKE 0 0 AAAQQQESATTQKAEKEVTRMVIIMVIFFLICWLPYASVAFYIFTHQGSNFGPIFMTLPAFFAKSSSIYNPVIYIMLNKQ 0 0 FRNCMLTTLCCGKNPLGDDDASATASKTETSQVAPA* 0 >MEL1_nanEhr Nannospalax ehrenbergi (molerat) OPN4 0 MNSPSGPRVPPGLAQKPSFMVTPVLPNQWISFQKNVSVGIQLPPASAT 0 0 ATGAQAASWVPFPTVDVPVHAHYTLGTVILLVGLTGMLGNLIVIYTFCR 2 1 SRGLRTRANMFTVNLAVSDFLMSFTQAPVFFASSLYKRWLFGEA 2 2 GCEFYAFCGAVSGITSMTTLTAIALDRYLVITRPLATIGVASKRRTALVLLGVWLYALAWSLPPFFGW 1 2 SAYVPEGLLTSCSWDYMTFTPSVRAYTMLLFCFVFFLPLLIIIFCYIFIFKAIRETGR 2 1 ACEGCGESPQRRRQWQRLQNEWKMAKVALLVIFLFVLSWAPYSTVALVAFAG 2 1 YSHILTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2 1 LAISQHLPCLGVLIGVSSQRSHPSLSYRSTHRSTLSSQASDLSWISGRKRQESLGSESEV 0 0 GWTDTEVTAAWGVAQEASGWSPYRHSLEDGEVKASPSPQGQEAKTSR 0 0 TKGQLPSLNLRM* 0 >PER_musMus Mus musculus (mouse) RRH peropsin syn(-CFI +NOLA1 +EGF -ELOVL6) 0 MLSEASDNSSGSRSEGSVFSRTEHSVIAAYLIVA 1 2 GITSILSNVVVLGIFIKYKELRTPTNAVIINLAFTDIGVSSIGYPMSAASDLHGSWKFGHAGCQ 0 0 IYAGLNIFFGMVSIGLLTVVAMDRYLTISCPDV 1 2 GRRMTTNTYLSMILGAWINGLFWALMPIIGWASYAPDPTGATCTINWRNNDT 2 1 SFVSYTMMVIVVNFIVPLTVMFYCYYHVSRSLRLYAASDCTAHLHRDWADQADVTK 0 0 MSVIMILMFLLAWSPYSIVCLWACFGNPKKIPPSMAIIAPLFAKSSTFYNPCIYVAAHKK 2 1 FRKAMLAMFKCQPHLAVPEPSTLPMDMPQSSLAPVRI* 0 >NEUR1_musMus Mus musculus (mouse) OPN5 neuropsin 0 MALNHTALPQDERLPHYLRDEDPFASKLSWEADLVAGFYLTII 1 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 2 VVGKPFTIISCFCHRWVFGWFGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 GVWLKRKHAYICLAVIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASGGGQVFILSILFFCLLLPTAVIVFSYAKIIAKVKSSSKEVAHFDSRIHSSHVLEVKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYRFACCQAGGLRGTKKKSLEDFR 2 1 LHTVTTVRKSSAVLEIHQEV* 0 >RGR_musMus Mus musculus (mouse) RGR syn(+PCDH21 -LRIT1 -GRID1 -WAPAL) 0 MAATRALPAGLGELEVLAVGTVLLME 1 2 ALSGISLNGLTIFSFCKTPDLRTPSNLLVLSLALADTGISLNALVAAVSSLLR 2 1 RWPHGSEGCQVHGFQGFATALASICGSAAVAWGRYHHYCT 1 2 GRQLAWDTAIPLVLFVWMSSAFWASLPLMGWGHYDYEPVGTCCTLDYSRGDR 2 1 NFISFLFTMAFFNFLVPLFITHTSYRFMEQKFSRSGHLP 0 0 VNTTLPGRMLLLGWGPYALLYLYAAIADVSFISPKLQM 0 0 VPALIAKTMPTINAINYALHREMVCRGTWQCLSPQKSKKDRTQ* 0