Uploads by Hiram
From genomewiki
Jump to navigationJump to search
This special page shows all uploaded files.
| Date | Name | Thumbnail | Size | Description | Versions |
|---|---|---|---|---|---|
| 02:56, 29 June 2012 | FireCracker10KElevationProfile.png (file) |  |
6 KB | Elevation profile for the Santa Cruz FireCracker 10K run, July 4th. Corrected the GPS readings with Google Earth information and known ground truth. | 2 |
| 22:05, 17 December 2008 | Flicek 11 DEC 2008.pdf (file) | 3.41 MB | Paul Flicek, EBI, presenting 1,000 genomes project status | 1 | |
| 17:42, 19 December 2008 | FrankJacobs181208.ppt (file) | 29.16 MB | Frank Jacobs from Rudolf Magnus Institute of Neuroscience, University of Utrecht, The Netherlands, "Pitx3 outSMRTs Nurr1 in dopamine neuron terminal differentiation" | 1 | |
| 17:33, 30 January 2019 | GalGal5.grcIncidentDb.bb (file) | 27 KB | gwUploadFile upload | 7 | |
| 17:24, 3 September 2019 | GalGal6.grcIncidentDb.bb (file) | 19 KB | gwUploadFile upload | 1 | |
| 22:10, 28 September 2017 | GenbankPrimates.jpg (file) | 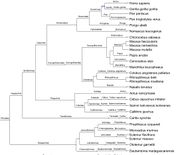 |
113 KB | primate phylogenetic tree from genbank taxIds Given a set of taxon IDs, utilizing [http://phylot.biobyte.de/ phylot.biobyte.de] to derive this phylogenetic tree. NOTE: not necessarily completely accurate. Image drawn by [http://itol.embl.de/ itol.embl. | 1 |
| 20:46, 14 October 2009 | Genecats 2009 10 14.ppt (file) | 141 KB | Building the kent source tree and a UCSC genome browser assembly, Genecats 2009-10-14 Hiram Clawson | 1 | |
| 20:46, 14 October 2009 | Genecats 2009 10 14.pptx (file) | 86 KB | Building the kent source tree and a UCSC genome browser assembly, Genecats 2009-10-14 Hiram Clawson | 1 | |
| 18:19, 19 March 2010 | Hg17.UCSC GGM BalancedCGIs.bb (file) | 389 KB | bigBed data for hg17 custom track: CGIs (balanced) Bona fide CpG islands, predicted to exhibit a combined epigenetic score above 0.5 | 1 | |
| 18:21, 19 March 2010 | Hg17.UCSC GGM CombinedEpigeneticScore.bb (file) | 2.02 MB | bigBed data for hg17 custom track: Epigenetic score - Combined epigenetic score (high scores correspond to bona fide CpG islands) | 1 | |
| 18:22, 19 March 2010 | Hg17.UCSC GGM OptimizedScore.bb (file) | 1.99 MB | bigBed data for hg17 custom track: Optimized score - Optimized score (high scores correspond to bona fide CpG islands | 1 | |
| 18:23, 19 March 2010 | Hg17.UCSC GGM SensitiveCGIs.bb (file) | 686 KB | bigBed data for hg17 custom track: CGIs (sensitive) - Bona fide CpG islands, predicted to exhibit a combined epigenetic score above 0.33 | 1 | |
| 18:25, 19 March 2010 | Hg17.UCSC GGM SpecificCGIs.bb (file) | 178 KB | bigBed data for hg17 custom track: CGIs (specific) - Bona fide CpG islands, predicted to exhibit a combined epigenetic score above 0.67 | 1 | |
| 18:27, 19 March 2010 | Hg17.customText.UCSC GGM All.txt (file) | 1 KB | customText track definition for all five data sets UCSC GGM CpGs | 1 | |
| 18:28, 19 March 2010 | Hg17.customText.UCSC GGM BalancedCGIs.txt (file) | 257 bytes | customText track definition for hg17 custom track: CGIs (balanced) - Bona fide CpG islands, predicted to exhibit a combined epigenetic score above 0.5 | 1 | |
| 18:29, 19 March 2010 | Hg17.customText.UCSC GGM CombinedEpigeneticScore.txt (file) | 272 bytes | customText track definition for hg17 custom track: Epigenetic score - Combined epigenetic score (high scores correspond to bona fide CpG islands) | 1 | |
| 18:30, 19 March 2010 | Hg17.customText.UCSC GGM SensitiveCGIs.txt (file) | 296 bytes | customText track definition for hg17 custom track: CGIs (sensitive) - Bona fide CpG islands, predicted to exhibit a combined epigenetic score above 0.33 | 1 | |
| 18:31, 19 March 2010 | Hg17.customText.UCSC GGM SpecificCGIs.txt (file) | 257 bytes | customText track definition for hg17 custom track: CGIs (specific) - Bona fide CpG islands, predicted to exhibit a combined epigenetic score above 0.67 | 1 | |
| 16:43, 18 September 2007 | Hg17 hg18.exonCount.png (file) |  |
5 KB | Human hg17 and hg18, exon count by size histogram, to size 30 nucleotides | 2 |
| 16:45, 18 September 2007 | Hg17 hg18.exonSize.png (file) |  |
5 KB | Human hg17 and hg18 exon size histogram, to size 50 nucleotides | 2 |
| 16:46, 18 September 2007 | Hg17 hg18.intronSize.png (file) |  |
5 KB | Human hg17 and hg18 intron size histogram to size 50 nucleotides | 2 |
| 16:46, 18 September 2007 | Hg17 hg18.intronsTo170.png (file) |  |
5 KB | Human hg17 and hg18 intron size to 170 nucleotides | 2 |
| 16:47, 18 September 2007 | Hg17 hg18 exonsTo300.png (file) |  |
6 KB | Human hg17 and hg18 exon size histogram, to a size of 300 nucleotides | 2 |
| 17:27, 19 March 2010 | Hg18.UCSC GGM BalancedCGIs.bb (file) | 389 KB | bigBed data for custom track of Bona fide CpG islands, predicted to exhibit a combined epigenetic score above 0.5 | 1 | |
| 17:28, 19 March 2010 | Hg18.UCSC GGM CombinedEpigeneticScore.bb (file) | 2.02 MB | bigBed data for hg18 custom track: Combined epigenetic score (high scores correspond to bona fide CpG islands) | 1 | |
| 17:28, 19 March 2010 | Hg18.UCSC GGM OptimizedScore.bb (file) | 1.99 MB | bigBed data for hg18 custom track: Optimized score (high scores correspond to bona fide CpG islands) | 1 | |
| 17:30, 19 March 2010 | Hg18.UCSC GGM SensitiveCGIs.bb (file) | 686 KB | bigBed data for custom track: Bona fide CpG islands, predicted to exhibit a combined epigenetic score above 0.33 | 1 | |
| 17:30, 19 March 2010 | Hg18.UCSC GGM SpecificCGIs.bb (file) | 178 KB | bigBed data for hg18 custom track: Bona fide CpG islands, predicted to exhibit a combined epigenetic score above 0.67 | 1 | |
| 17:54, 19 March 2010 | Hg18.customText.UCSC GGM All.txt (file) | 1 KB | customText track statements to load all five UCSC_GGM CpG custom tracks | 2 | |
| 18:00, 19 March 2010 | Hg18.customText.UCSC GGM BalancedCGIs.txt (file) | 257 bytes | customText track definition for hg18 custom track: Bona fide CpG islands, predicted to exhibit a combined epigenetic score above 0.5 | 1 | |
| 18:07, 19 March 2010 | Hg18.customText.UCSC GGM CombinedEpigeneticScore.txt (file) | 272 bytes | customText for hg18 custom track: Combined epigenetic score (high scores correspond to bona fide CpG islands) | 1 | |
| 18:06, 19 March 2010 | Hg18.customText.UCSC GGM SensitiveCGIs.txt (file) | 259 bytes | customText for hg18 custom track: Bona fide CpG islands, predicted to exhibit a combined epigenetic score above 0.33 | 1 | |
| 18:05, 19 March 2010 | Hg18.customText.UCSC GGM SpecificCGIs.txt (file) | 257 bytes | customText for hg18 custom track: Bona fide CpG islands, predicted to exhibit a combined epigenetic score above 0.67 | 1 | |
| 23:29, 18 September 2007 | Hg18KnownGenesSmallIntronsExonsCt.txt.gz (file) | 122 KB | Human Hg18 genome browser custom track to indicate unusually small introns and exons in the Known Genes version 3 procedure. These small exons and introns are used to maintain frame shifts in mRNA evidence that may be translation skipping or post translat | 1 | |
| 16:33, 28 June 2022 | Hg19.grcIncidentDb.bb (file) | 106 KB | gwUploadFile upload | 867 | |
| 16:43, 27 April 2012 | Hg19.ncbiIncidentDb.bb (file) | 179 KB | gwUploadFile upload | 67 | |
| 18:46, 15 September 2022 | Hg38.grcIncidentDb.bb (file) | 282 KB | gwUploadFile upload | 359 | |
| 18:39, 16 December 2008 | ISMB2007TripReportGenecats.ppt (file) | 4.42 MB | Rachel Harte genecats presentation summary from ISMB 2007, Vienna Austria | 1 | |
| 14:39, 23 July 2008 | ISMB2008 UCSC.ppt (file) | 5.35 MB | ISMB 2008 Toronto - Intelligent Systems for Molecular Biology - presentation by Hiram Clawson. An example table browser intersection is performed with all the steps indicated. | 1 | |
| 22:00, 6 August 2008 | ISMB2008 genecats summary.ppt (file) | 3.8 MB | Hiram's summary of ISMB 2008 Toronto Ontario, July 2008 | 1 | |
| 17:10, 26 September 2018 | InitParasol.sh.txt (file) | 3 KB | less restrictive subnet specification and separate hub/node logs in ./logs/ directory | 3 | |
| 18:57, 16 December 2008 | Kent allJoiner.ppt (file) | 41 KB | Jim Kent, genecats 05 July 2008 - the all.joiner file which describes the database schema relationships for the UCSC genome browser | 1 | |
| 18:44, 17 December 2008 | Kent decodingEncode.ppt (file) | 4.24 MB | Jim Kent at the Computational Systems Bioinformatics Conference, Stanford CA, 27 Aug 2008. Encode, the genome browser, the table browser, the source code, a variety of subjects are discussed. | 1 | |
| 19:02, 16 December 2008 | Kent parasolArchitecture.ppt (file) | 60 KB | Jim Kent genecats presentation June 2008, Parasol super computer job management system review | 1 | |
| 18:04, 15 September 2010 | Knetfile hooks.0.1.7.patch (file) | 13 KB | Patch to samtools-0.1.7 to enable UCSC network functions and caching for remote BAM file access | 1 | |
| 18:03, 15 September 2010 | Knetfile hooks.0.1.8.patch (file) | 16 KB | Patch to samtools-0.1.8 to enable UCSC network functions and caching for remote BAM file access | 1 | |
| 17:09, 28 April 2018 | LastzProcessingTimeHistogram.png (file) |  |
53 KB | lastz cluster compute time histogram 1,635 measurements from alignments performed at UCSC 459 measurements are less than 0.05 days (less than 1 hour) due to very small genome sizes 59 measurements are longer than 5 days processing time due to high c... | 1 |
| 00:11, 15 October 2022 | Mac background Catalina.png (file) |  |
6.2 MB | test upload function and thumbnail | 1 |
| 23:18, 18 March 2015 | MafScoreSizeScan pl.txt (file) | 2 KB | script used to scan a MAF file to extract scores, sizes, and coverage statistics of each alignment block. Used in lastz tuning procedures. | 1 | |
| 17:43, 19 March 2015 | MatrixSummary pl.txt (file) | 6 KB | used in lastz tuning procedure, examines output of the four lastz_D outputs to survey the differences | 1 |