Uploads by Hiram
From genomewiki
Jump to navigationJump to search
This special page shows all uploaded files.
| Date | Name | Thumbnail | Size | Description | Versions |
|---|---|---|---|---|---|
| 19:09, 22 January 2026 | BME-110.2026-01-22.pptx (file) | 13.15 MB | BME 110 Browser introduction 22 Jan 2026 | 1 | |
| 19:07, 22 January 2026 | BIOL-104A.2026-01-27.pptx (file) | 13.15 MB | Biology 104A - 27 January 2026 | 1 | |
| 19:35, 13 January 2026 | PAG 2026.pptx (file) | 6.85 MB | PAG 2026 - San Diego - Monday 12 January 2026 | 1 | |
| 17:17, 21 November 2025 | VGP 2025 RockefellerU.jpg (file) |  |
2 MB | VGP 2025 meeting group photograph, Rockefeller University, New York | 1 |
| 05:46, 1 November 2025 | BME18-05Nov2025.pptx (file) | 12.41 MB | BME 18 presentation 05 Nov 2025 | 1 | |
| 17:20, 4 August 2025 | FindGenome.txt (file) | 3 KB | 1 | ||
| 22:43, 15 April 2025 | BME-110.2025-04-17.pptx (file) | 13.12 MB | BME 110 browser presentation, 17 April 2025 | 1 | |
| 22:47, 2 April 2025 | RIKEN 2025.pptx (file) | 4.84 MB | RIKEN Yokohama, 2025 - recent updates to the UCSC genome browser | 1 | |
| 04:20, 3 February 2025 | BIOL-104A.2025-02-05.pptx (file) | 13.12 MB | Biology 104A Human Genetic Variation Lab I | 1 | |
| 20:08, 21 January 2025 | BME-110.2025-01-23.pptx (file) | 13.12 MB | BME 110 class presentation, introduction to the genome browser | 1 | |
| 17:20, 11 January 2025 | Sat Workshop2025.pptx (file) | 5.54 MB | PAG San Diego 2025, Hiram Clawson - Finding Genomes in the Genome Browser | 1 | |
| 17:54, 28 October 2024 | BME-18.2024-10-30.pptx (file) | 12.27 MB | Updated slides adding extra topics - BME 18 Wed. 2024-10-30 | 1 | |
| 21:59, 17 October 2024 | Vgp 2024-10-18.pptx (file) | 4.16 MB | curl -I test on actual GenomeArk file | 1 | |
| 02:21, 22 April 2024 | BME-110.2024-04-25.pptx (file) | 12.97 MB | BME 110 25 April 2024 - UCSC browser introduction | 1 | |
| 21:04, 18 January 2024 | BME-110.2024-01-18.pptx (file) | 12.97 MB | 1 | ||
| 22:32, 8 January 2024 | CSHL GenomeInformatics 2023.pptx (file) | 4.9 MB | Cold Spring Harbor - Genome Informatics meeting - poster - GenArk assembly resources | 1 | |
| 22:18, 8 January 2024 | Sat Workshop2024.pptx (file) | 4.45 MB | PAG 31 - 2024 - San Diego Plant and Animal Genomes - GenArk assembly resources | 1 | |
| 18:52, 3 November 2023 | BME-18.2023-11-08.pptx (file) | 8.83 MB | 1 | ||
| 21:01, 6 October 2023 | BME-110.2023-10-06.pptx (file) | 9.49 MB | 2 | ||
| 17:05, 15 January 2023 | PAG30 2023-01-15.pptx (file) | 7.55 MB | presentation at the 30th annual Plant and Animal Genomes conference - "Educational Resources in the UCSC Genome Browser" - introducing the http://genome.ucsc.edu/training/education/ resources. | 1 | |
| 17:00, 15 January 2023 | PAG30 2023-01-14.pptx (file) | 9.85 MB | presentation at the Plant And Animal Genomes conference, San Diego January 2023 - "The UCSC Genome Browser" - an introduction to the browser, GenArk assembly hubs, building your own assembly hub and REST API interface. | 1 | |
| 18:56, 4 November 2022 | BME-18.2022-11-04.pptx (file) | 8.59 MB | 1 | ||
| 00:11, 15 October 2022 | Mac background Catalina.png (file) |  |
6.2 MB | test upload function and thumbnail | 1 |
| 18:46, 15 September 2022 | Hg38.grcIncidentDb.bb (file) | 282 KB | gwUploadFile upload | 359 | |
| 16:33, 28 June 2022 | DanRer11.grcIncidentDb.bb (file) | 202 KB | gwUploadFile upload | 17 | |
| 16:33, 28 June 2022 | DanRer10.grcIncidentDb.bb (file) | 126 KB | gwUploadFile upload | 127 | |
| 16:33, 28 June 2022 | Mm9.grcIncidentDb.bb (file) | 74 KB | gwUploadFile upload | 282 | |
| 16:33, 28 June 2022 | DanRer7.grcIncidentDb.bb (file) | 111 KB | gwUploadFile upload | 336 | |
| 16:33, 28 June 2022 | Mm10.grcIncidentDb.bb (file) | 101 KB | gwUploadFile upload | 289 | |
| 16:33, 28 June 2022 | Hg19.grcIncidentDb.bb (file) | 106 KB | gwUploadFile upload | 867 | |
| 22:32, 20 May 2021 | VGP 2021-05-21.pptx (file) | 1.24 MB | VGP meeting 2021-05-21 presentation, VGP assembly resources in the UCSC genome browser | 1 | |
| 17:24, 3 September 2019 | GalGal6.grcIncidentDb.bb (file) | 19 KB | gwUploadFile upload | 1 | |
| 00:56, 19 May 2019 | NationalYoYoMuseum ChicoCA 50.jpg (file) |  |
553 KB | National YO YO Museum, Chico CA | 1 |
| 17:33, 30 January 2019 | GalGal5.grcIncidentDb.bb (file) | 27 KB | gwUploadFile upload | 7 | |
| 21:54, 1 October 2018 | AwsAttachVolume.sh.txt (file) | 4 KB | Script for AWS instance initialization to attach a data volume to a running instance. Used in the startParaHub.sh script upon starting the machine. | 1 | |
| 17:35, 26 September 2018 | OpenStackParaHubSetup.sh.txt (file) | 8 KB | reorganize NFS setups, parameterize the installation directory | 5 | |
| 17:10, 26 September 2018 | InitParasol.sh.txt (file) | 3 KB | less restrictive subnet specification and separate hub/node logs in ./logs/ directory | 3 | |
| 20:32, 25 September 2018 | NodeReport.sh.txt (file) | 1 KB | add usage message and directory argument for result file | 4 | |
| 17:42, 11 July 2018 | OpenStackParaNodeSetup.sh.template.txt (file) | 4 KB | correctly fixup /mnt filesystem for broken openstack installation, and mount /mnt/data from the hub | 2 | |
| 17:09, 28 April 2018 | LastzProcessingTimeHistogram.png (file) |  |
53 KB | lastz cluster compute time histogram 1,635 measurements from alignments performed at UCSC 459 measurements are less than 0.05 days (less than 1 hour) due to very small genome sizes 59 measurements are longer than 5 days processing time due to high c... | 1 |
| 23:13, 27 April 2018 | SizeVsTime.png (file) |  |
156 KB | lastz cluster run processing time vs. genome size blue points plot times 2 days or less processing time, left axis scale 0 - 2 days red points plot times over 2 days processing time, right axis scale 0 - 70 days | 1 |
| 02:53, 6 April 2018 | ParasolInstall.sh.txt (file) | 1 KB | adding git fetch of automation scripts into /data/scripts/ | 3 | |
| 22:10, 28 September 2017 | GenbankPrimates.jpg (file) | 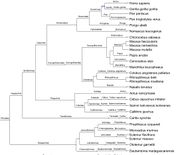 |
113 KB | primate phylogenetic tree from genbank taxIds Given a set of taxon IDs, utilizing [http://phylot.biobyte.de/ phylot.biobyte.de] to derive this phylogenetic tree. NOTE: not necessarily completely accurate. Image drawn by [http://itol.embl.de/ itol.embl. | 1 |
| 18:17, 2 June 2017 | Btrfs filesystem readWrite performance.png (file) |  |
120 KB | read/write performance test results for 'btrfs' filesystem | 1 |
| 22:48, 31 May 2017 | Xfs filesystem readWrite performance.png (file) |  |
97 KB | testing read/write performance on xfs filesystem in a directory overloaded with up to 7 million files | 1 |
| 22:47, 31 May 2017 | Tmpfs filesystem readWrite performance.png (file) |  |
101 KB | testing read/write performance into a /dev/shm directory overloaded with up to 7 million files | 1 |
| 22:45, 31 May 2017 | Ext4 fs readWrite performance.png (file) |  |
106 KB | writing over 7 million files into one directory to see how performance degrades. | 1 |
| 18:26, 26 January 2017 | BME230 Winter 2017.ppt (file) | 12.98 MB | (Hiram's presentation to BME230 Winter 2016 12 January - brief browser usage introduction, assembly hub demonstration, table browser intersection, plus information on source code but not part of the discussion) | 1 | |
| 05:39, 9 June 2016 | BejeranoLab 2016-06-09.pptx (file) | 2.92 MB | 2 | ||
| 18:22, 12 January 2016 | BME230 Winter 2016.ppt (file) | 13.8 MB | (Hiram's presentation to BME230 Winter 2016 12 January - brief browser usage introduction, assembly hub demonstration, table browser intersection, plus information on source code but not part of the discussion)) | 1 |